- Course overview
- Search within this course
- What is UniProt?
- Where does the data come from?
- Why do we need UniProt?
- When to use UniProt
- Quiz: Check your learning I
- How to access and navigate UniProt
- How to search UniProt
- Annotation score
- Quiz: Check your learning II
- Exploring a UniProtKB entry
- How to get data from UniProt
- How to submit data to UniProt
- When to use UniProt: guided examples
- Exercise: finding entries with 3D structures
- Exercise: mapping other database identifiers to UniProt
- Summary
- Get help and support on UniProt
- References
- Next steps
- Your feedback
How to use UniProt tools
What analysis tools are provided?
UniProt provides four tools to analyse protein data. These are: 1) Blast; 2) Align; 3) ID mapping; 4) Peptide search (Figure 46).
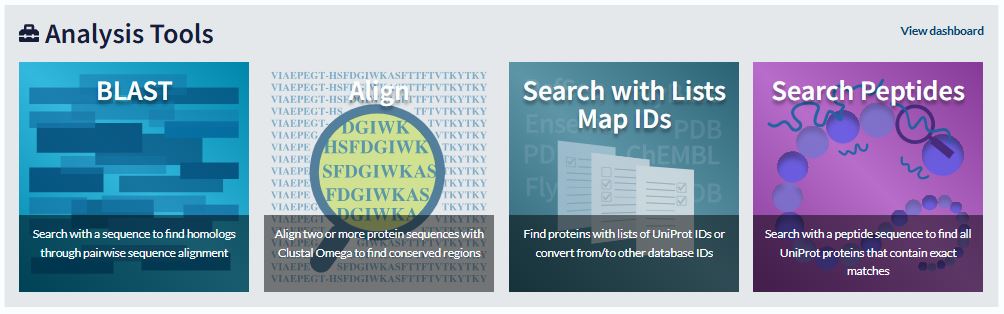
What do the tools do?
1) ‘BLAST’ (Basic Local Alignment Search Tool) is used for sequence similarity searching.
2) ‘Align’ is used for multiple sequence alignment.
3) ‘Peptide search’ allows you to submit short peptide sequences of at least three residues and find all UniProtKB sequences which have an exact match to the query sequence.
4) ‘ID mapping’ allows you to use a list of identifiers to retrieve batches of UniProtKB entries and to convert database identifiers from UniProt to external databases or vice versa.
How to access the tools
Header links: All four analysis tools can be accessed from links in the header which appears at the top of each page (Figure 47).

Tools tab: You can also access the BLAST, Align and ID mapping tools from the ‘Tools’ tab in each entry (Figure 48).
