- Course overview
- Search within this course
- What is UniProt?
- Where does the data come from?
- Why do we need UniProt?
- When to use UniProt
- Quiz: Check your learning I
- How to access and navigate UniProt
- How to search UniProt
- Annotation score
- Quiz: Check your learning II
- How to use UniProt tools
- How to get data from UniProt
- How to submit data to UniProt
- When to use UniProt: guided examples
- Exercise: finding entries with 3D structures
- Exercise: mapping other database identifiers to UniProt
- Summary
- Get help and support on UniProt
- References
- Next steps
- Your feedback
Function
This section of a UniProt entry provides a wide range of information about the role of the protein in a cell, including a general description of its function which, in the case of an enzyme, includes a description of its catalytic activity, required cofactors and factors which regulate its activity (Figure 24).

Gene Ontology (GO) terms are also provided. GO is a controlled vocabulary which provides an overview of the functions and processes in which a protein is involved as well as where it is located in the cell. GO annotations are provided in two different tabs. The ‘GO annotations’ tab is displayed by default (Figure 25a). It shows a graphical tool which provides a quick visual summary by grouping the GO annotations into high level terms selected to represent a broad range of functions. Below this, a table lists the terms with one annotation per row.

A second tab, ‘GO-CAM models’, displays GO Causal Activity Models when available (Figure 25b). GO-CAMs depict how proteins interact and function within biological systems.

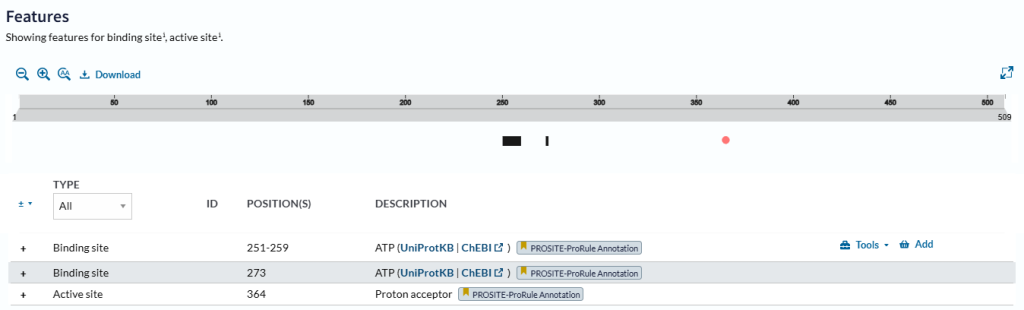
Functional sites in the protein sequence such as binding sites and active sites are shown, both graphically and in an embedded viewer (Figure 26).