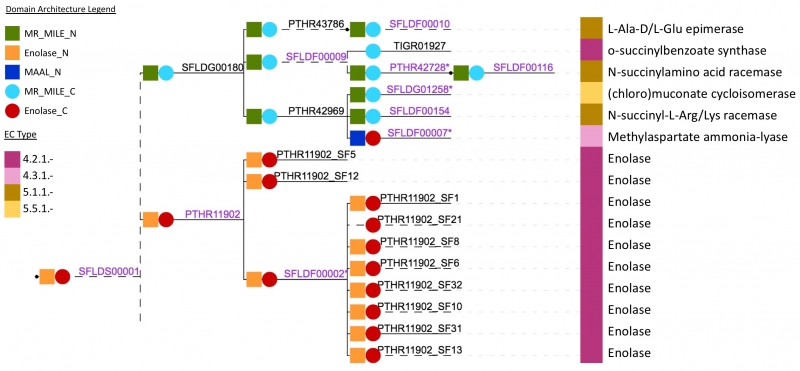
Predicting protein signature relationships
It is possible to automatically predict relationships between signatures by calculating the Jaccard index (a statistic used for comparing the similarity of sample sets, in this case protein matches) for each pair of signatures. This allows us to produce a predicted tree of relationships. An example of this semi-automatically generated tree is shown below (Figure 3) for a subset of the SFLD superfamily ‘Enolase’. While this system functions as a useful preliminary guide for InterPro curators, manual curation of most relationships between signatures is still required.