 |
PDBsum entry 6d4t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
6d4t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.205
- Imp dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NAD+ + H2O = XMP + NADH + H+
|
 |
 |
 |
 |
 |
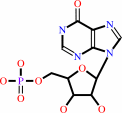 IMP
IMP
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
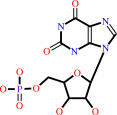 XMP
XMP
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Eur J Med Chem
174:309-329
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
|
|
V.Singh,
A.Pacitto,
S.Donini,
D.M.Ferraris,
S.Boros,
E.Illyés,
B.Szokol,
M.Rizzi,
T.L.Blundell,
D.B.Ascher,
J.Pato,
V.Mizrahi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tuberculosis (TB) is a major infectious disease associated increasingly with
drug resistance. Thus, new anti-tubercular agents with novel mechanisms of
action are urgently required for the treatment of drug-resistant TB. In prior
work, we identified compound 1
(cyclohexyl(4-(isoquinolin-5-ylsulfonyl)piperazin-1-yl)methanone) and showed
that its anti-tubercular activity is attributable to inhibition of
inosine-5'-monophosphate dehydrogenase (IMPDH) in Mycobacterium tuberculosis. In
the present study, we explored the structure-activity relationship around
compound 1 by synthesizing and evaluating the inhibitory activity of analogues
against M. tuberculosis IMPDH in biochemical and whole-cell assays. X-ray
crystallography was performed to elucidate the mode of binding of selected
analogues to IMPDH. We establish the importance of the cyclohexyl, piperazine
and isoquinoline rings for activity, and report the identification of an
analogue with IMPDH-selective activity against a mutant of M. tuberculosis that
is highly resistant to compound 1. We also show that the nitrogen in urea
analogues is required for anti-tubercular activity and identify benzylurea
derivatives as promising inhibitors that warrant further investigation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

