 |
PDBsum entry 4xkm
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of xylose isomerase from an human intestinal tract microbe bacteroides thetaiotaomicron
|
|
Structure:
|
 |
Xylose isomerase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482). Organism_taxid: 226186. Strain: atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482. Gene: xyla, bt_0793. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.187
|
R-free:
|
0.211
|
|
|
Authors:
|
 |
B.G.Han,S.M.Bong,J.W.Cho,B.I.Lee
|
|
Key ref:
|
 |
B.G.Han
et al.
(2015).
Crystal structure of a class 2 d-Xylose isomerase fro human intestinal tract microbe bacteroides thetaiotao.
Biodesign,
3,
41.
|
 |
|
Date:
|
 |
|
12-Jan-15
|
Release date:
|
23-Dec-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8A9M2
(XYLA_BACTN) -
Xylose isomerase from Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
438 a.a.
435 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.5
- xylose isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-xylose = alpha-D-xylulofuranose
|
 |
 |
 |
 |
 |
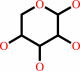 alpha-D-xylose
alpha-D-xylose
|
=
|
alpha-D-xylulofuranose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

