 |
PDBsum entry 4pv1

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Electron transport/inhibitor
|
PDB id
|
|

|

|
4pv1
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 213 a.a.
213 a.a.
|
 |

|

|

|

|

|

|

|
 159 a.a.
159 a.a.
|
 |

|

|

|

|

|

|

|
 288 a.a.
288 a.a.
|
 |

|

|

|

|

|

|

|
 158 a.a.
158 a.a.
|
 |

|

|

|

|

|

|

|
 28 a.a.
28 a.a.
|
 |

|

|

|

|

|

|

|
 31 a.a.
31 a.a.
|
 |

|

|

|

|

|

|

|
 37 a.a.
37 a.a.
|
 |

|

|

|

|

|

|

|
 28 a.a.
28 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Electron transport/inhibitor
|
 |
|
Title:
|
 |
Cytochrome b6f structure from m. Laminosus with the quinone analog inhibitor stigmatellin
|
|
Structure:
|
 |
Cytochrome b6. Chain: a. Cytochrome b6-f complex subunit 4. Chain: b. Synonym: 17 kda polypeptide. Apocytochrome f. Chain: c. Fragment: unp residues 45-333. Cytochrome b6-f complex iron-sulfur subunit.
|
|
Source:
|
 |
Mastigocladus laminosus. Organism_taxid: 83541. Organism_taxid: 83541
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.217
|
R-free:
|
0.247
|
|
|
Authors:
|
 |
S.S.Hasan,E.Yamashita,W.A.Cramer
|
|
Key ref:
|
 |
S.S.Hasan
et al.
(2014).
Traffic within the cytochrome b6f lipoprotein complex: gating of the quinone portal.
Biophys J,
107,
1620-1628.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
14-Mar-14
|
Release date:
|
20-Aug-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P83791
(CYB6_MASLA) -
Cytochrome b6 from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
215 a.a.
213 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P83792
(PETD_MASLA) -
Cytochrome b6-f complex subunit 4 from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
160 a.a.
159 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P83793
(CYF_MASLA) -
Cytochrome f from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
333 a.a.
288 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P83794
(UCRI_MASLA) -
Cytochrome b6-f complex iron-sulfur subunit from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
179 a.a.
158 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P83795
(PETL_MASLA) -
Cytochrome b6-f complex subunit 6 from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
32 a.a.
28 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P83796
(PETM_MASLA) -
Cytochrome b6-f complex subunit 7 from Mastigocladus laminosus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
35 a.a.
31 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain D:
E.C.7.1.1.6
- plastoquinol--plastocyanin reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 oxidized [plastocyanin] + a plastoquinol + 2 H+(in) = 2 reduced [plastocyanin] + a plastoquinone + 4 H+(out)
|
 |
 |
 |
 |
 |
2
×
oxidized [plastocyanin]
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
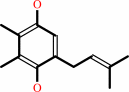 plastoquinol
plastoquinol
|
+
|
2
×
H(+)(in)
|
=
|
2
×
reduced [plastocyanin]
|
+
|
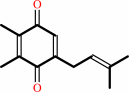 plastoquinone
plastoquinone
|
+
|
4
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biophys J
107:1620-1628
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Traffic within the cytochrome b6f lipoprotein complex: gating of the quinone portal.
|
|
S.S.Hasan,
E.A.Proctor,
E.Yamashita,
N.V.Dokholyan,
W.A.Cramer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The cytochrome bc complexes b6f and bc1 catalyze proton-coupled quinol/quinone
redox reactions to generate a transmembrane proton electrochemical gradient.
Quinol oxidation on the electrochemically positive (p) interface of the complex
occurs at the end of a narrow quinol/quinone entry/exit Qp portal, 11 Å long in
bc complexes. Superoxide, which has multiple signaling functions, is a
by-product of the p-side quinol oxidation. Although the transmembrane core and
the chemistry of quinone redox reactions are conserved in bc complexes, the rate
of superoxide generation is an order of magnitude greater in the b6f complex,
implying that functionally significant differences in structure exist between
the b6f and bc1 complexes on the p-side. A unique structure feature of the b6f
p-side quinol oxidation site is the presence of a single chlorophyll-a molecule
whose function is unrelated to light harvesting. This study describes a
cocrystal structure of the cytochrome b6f complex with the quinol analog
stigmatellin, which partitions in the Qp portal of the bc1 complex, but not
effectively in b6f. It is inferred that the Qp portal is partially occluded in
the b6f complex relative to bc1. Based on a discrete molecular-dynamics
analysis, occlusion of the Qp portal is attributed to the presence of the
chlorophyll phytyl tail, which increases the quinone residence time within the
Qp portal and is inferred to be a cause of enhanced superoxide production. This
study attributes a novel (to our knowledge), structure-linked function to the
otherwise enigmatic chlorophyll-a in the b6f complex, which may also be relevant
to intracellular redox signaling.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

