 |
PDBsum entry 3h9c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Structure of methionyl-tRNA synthetase: crystal form 2
|
|
Structure:
|
 |
Methionyl-tRNA synthetase. Chain: a. Fragment: m547 domain: unp residues 2-548. Synonym: methionine-tRNA ligase, metrs. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 83333. Strain: k-12. Gene: b2114, jw2101, metg. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.40Å
|
R-factor:
|
0.167
|
R-free:
|
0.191
|
|
|
Authors:
|
 |
E.Schmitt,I.C.Tanrikulu,T.H.Yoo,M.Panvert,D.A.Tirrell,Y.Mechulam
|
Key ref:

|
 |
E.Schmitt
et al.
(2009).
Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J Mol Biol,
394,
843-851.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
30-Apr-09
|
Release date:
|
08-Dec-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00959
(SYM_ECOLI) -
Methionine--tRNA ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
677 a.a.
544 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
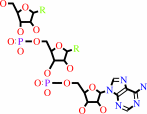 tRNA(Met)
tRNA(Met)
|
+
|
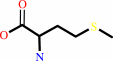 L-methionine
L-methionine
|
+
|
 ATP
ATP
|
=
|
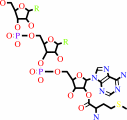 L-methionyl-tRNA(Met)
L-methionyl-tRNA(Met)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
394:843-851
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
|
|
E.Schmitt,
I.C.Tanrikulu,
T.H.Yoo,
M.Panvert,
D.A.Tirrell,
Y.Mechulam.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Methionyl-tRNA synthetase (MetRS) specifically binds its methionine substrate in
an induced-fit mechanism, with methionine binding causing large rearrangements.
Mutated MetRS able to efficiently aminoacylate the methionine (Met) analog
azidonorleucine (Anl) have been identified by saturation mutagenesis combined
with in vivo screening procedures. Here, the crystal structure of such a mutated
MetRS was determined in the apo form as well as complexed with Met or Anl (1.4
to 1.7 A resolution) to reveal the structural basis for the altered specificity.
The mutations result in both the loss of important contacts with Met and the
creation of new contacts with Anl, thereby explaining the specificity shift.
Surprisingly, the conformation induced by Met binding in wild-type MetRS already
occurs in the apo form of the mutant enzyme. Therefore, the mutations cause the
enzyme to switch from an induced-fit mechanism to a lock-and-key one, thereby
enhancing its catalytic efficiency.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Unliganded MetRS-SLL is in the met-on conformation.
Top panel: the active-site region of unliganded MetRS-SLL
(yellow) was superimposed to that of unliganded MetRS (PDB ID:
1QQT; blue). Motions of aromatic residues are indicated by
arrows. Bottom panel: same as top panel but unliganded MetRS was
replaced by MetRS:Met (PDB ID: 1F4L; blue). The bound methionine
is shown in green.
|
 |
Figure 4.
Fig. 4. Schematic representation of hydrophobic (red broken
lines) and electrostatic (green broken lines) interactions made
between MetRS-SLL and Anl. Anl is represented with blue bonds.
Carbons are in black, nitrogens are in dark blue, oxygens are in
red, and sulfurs are in yellow. Water molecules are symbolized
as light blue spheres. Relevant hydrogen-bonding distances are
indicated in angstroms. The electrostatic interaction between
Wat2 and the N2 atom of Anl is indicated by a blue broken line.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2009,
394,
843-851)
copyright 2009.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Ingvarsson,
and
T.Unge
(2010).
Flexibility and communication within the structure of the Mycobacterium smegmatis methionyl-tRNA synthetase.
|
| |
FEBS J,
277,
3947-3962.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

