 |
PDBsum entry 3a2b

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.3.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.3.1.47
- 8-amino-7-oxononanoate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
6-carboxyhexanoyl-[ACP] + L-alanine + H+ = (8S)-8-amino-7-oxononanoate + holo-[ACP] + CO2
|
 |
 |
 |
 |
 |
6-carboxyhexanoyl-[ACP]
|
+
|
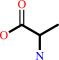 L-alanine
L-alanine
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
=
|
(8S)-8-amino-7-oxononanoate
|
+
|
holo-[ACP]
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.2.3.1.50
- serine C-palmitoyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-serine + hexadecanoyl-CoA + H+ = 3-oxosphinganine + CO2 + CoA
|
 |
 |
 |
 |
 |
 L-serine
L-serine
|
+
|
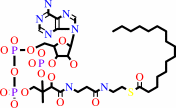 hexadecanoyl-CoA
hexadecanoyl-CoA
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
3-oxosphinganine
|
+
|
 CO2
CO2
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
146:549-562
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the enzymatic mechanism of serine palmitoyltransferase from Sphingobacterium multivorum.
|
|
H.Ikushiro,
M.M.Islam,
A.Okamoto,
J.Hoseki,
T.Murakawa,
S.Fujii,
I.Miyahara,
H.Hayashi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Serine palmitoyltransferase (SPT) is a key enzyme of sphingolipid biosynthesis
and catalyses the pyridoxal 5'-phosphate (PLP)-dependent decarboxylative
condensation reaction of l-serine with palmitoyl-CoA to generate
3-ketodihydrosphingosine. The crystal structure of SPT from Sphingobacterium
multivorum GTC97 complexed with l-serine was determined at 2.3 A resolution. The
electron density map showed the Schiff base formation between l-serine and PLP
in the crystal. Because of the hydrogen bond formation with His138, the
orientation of the Calpha-H bond of the PLP-l-serine aldimine was not
perpendicular to the PLP-Schiff base plane. This conformation is unfavourable
for the alpha-proton abstraction by Lys244 and the reaction is expected to stop
at the PLP-l-serine aldimine. Structural modelling of the following
intermediates indicated that His138 changes its hydrogen bond partner from the
carboxyl group of l-serine to the carbonyl group of palmitoyl-CoA upon the
binding of palmitoyl-CoA, making the l-serine Calpha-H bond perpendicular to the
PLP-Schiff base plane. These crystal and model structures well explained the
observations on bacterial SPTs that the alpha-deprotonation of l-serine occurs
only in the presence of palmitoyl-CoA. This study provides the structural
evidence that directly supports our proposed mechanism of the substrate
synergism in the SPT reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Bourquin,
H.Riezman,
G.Capitani,
and
M.G.Grütter
(2010).
Structure and function of sphingosine-1-phosphate lyase, a key enzyme of sphingolipid metabolism.
|
| |
Structure,
18,
1054-1065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Raman,
K.A.Johnson,
D.J.Clarke,
J.H.Naismith,
and
D.J.Campopiano
(2010).
The serine palmitoyltransferase from Sphingomonas wittichii RW1: An interesting link to an unusual acyl carrier protein.
|
| |
Biopolymers,
93,
811-822.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

