
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 436 a.a.
436 a.a.
|
 |

|

|

|

|

|

|

|

 380 a.a.
380 a.a.
|
 |

|

|

|

|

|

|

|

 103 a.a.
103 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The crystal structure of desulfovibrio vulgaris dissimilatory sulfite reductase bound to dsrc provides novel insights into the mechanism of sulfate respiration
|
|
Structure:
|
 |
Sulfite reductase, dissimilatory-type subunit alpha. Chain: a, d. Synonym: dissimilatory sulfite reductase, desulfoviridin subunit alpha, hydrogensulfite reductase alpha subunit. Sulfite reductase, dissimilatory-type subunit beta. Chain: b, e. Synonym: dissimilatory sulfite reductase, desulfoviridin subunit beta, hydrogensulfite reductase subunit beta. Sulfite reductase, dissimilatory-type subunit gamma.
|
|
Source:
|
 |
Desulfovibrio vulgaris. Organism_taxid: 882. Strain: hildenborough. Atcc: 29579. Atcc: 29579
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.192
|
R-free:
|
0.219
|
|
|
Authors:
|
 |
T.F.Oliveira,C.Vonrhein,P.M.Matias,S.S.Venceslau,I.A.C.Pereira, M.Archer
|
Key ref:

|
 |
T.F.Oliveira
et al.
(2008).
The Crystal Structure of Desulfovibrio vulgaris Dissimilatory Sulfite Reductase Bound to DsrC Provides Novel Insights into the Mechanism of Sulfate Respiration.
J Biol Chem,
283,
34141-34149.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
22-Sep-08
|
Release date:
|
02-Dec-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P45574
(DSVA_DESVH) -
Sulfite reductase, dissimilatory-type subunit alpha from Nitratidesulfovibrio vulgaris (strain ATCC 29579 / DSM 644 / CCUG 34227 / NCIMB 8303 / VKM B-1760 / Hildenborough)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
437 a.a.
436 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, D:
E.C.1.8.99.3
- Transferred entry: 1.8.99.5.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(O3S.S.SO(3))2- + acceptor + 2 H2O + OH- = 3 HSO(3)- + reduced acceptor
|
 |
 |
 |
 |
 |
 (O(3)S.S.SO(3))(2-)
(O(3)S.S.SO(3))(2-)
|
+
|
 acceptor
acceptor
|
+
|
2
×
H(2)O
|
+
|
OH(-)
|
=
|
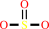 3
×
HSO(3)(-)
3
×
HSO(3)(-)
|
+
|
reduced acceptor
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur; Siroheme
|
 |
 |
 |
 |
 |
Iron-sulfur
Bound ligand (Het Group name =
SH0)
matches with 98.41% similarity
|
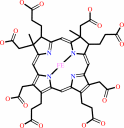 Siroheme
Siroheme
|
|
 |
 |
Enzyme class 3:
|
 |
Chains B, C, E, F:
E.C.1.8.1.22
- dissimilatory sulfite reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[DsrC protein]-trisulfide + NAD+ + 3 H2O = [DsrC protein]-dithiol + sulfite + NADH + 3 H+
|
 |
 |
 |
 |
 |
[DsrC protein]-trisulfide
|
+
|
 NAD(+)
NAD(+)
|
+
|
3
×
H2O
|
=
|
[DsrC protein]-dithiol
|
+
|
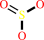 3
×
sulfite
3
×
sulfite
|
+
|
 NADH
NADH
|
+
|
3
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:34141-34149
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The Crystal Structure of Desulfovibrio vulgaris Dissimilatory Sulfite Reductase Bound to DsrC Provides Novel Insights into the Mechanism of Sulfate Respiration.
|
|
T.F.Oliveira,
C.Vonrhein,
P.M.Matias,
S.S.Venceslau,
I.A.Pereira,
M.Archer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Sulfate reduction is one of the earliest types of energy metabolism used by
ancestral organisms to sustain life. Despite extensive studies, many questions
remain about the way respiratory sulfate reduction is associated with energy
conservation. A crucial enzyme in this process is the dissimilatory sulfite
reductase (dSiR), which contains a unique siroheme-[4Fe4S] coupled cofactor.
Here, we report the structure of desulfoviridin from Desulfovibrio vulgaris, in
which the dSiR DsrAB (sulfite reductase) subunits are bound to the DsrC protein.
The alpha(2)beta(2)gamma(2) assembly contains two siroheme-[4Fe4S] cofactors
bound by DsrB, two sirohydrochlorins and two [4Fe4S] centers bound by DsrA, and
another four [4Fe4S] centers in the ferredoxin domains. A sulfite molecule,
coordinating the siroheme, is found at the active site. The DsrC protein is
bound in a cleft between DsrA and DsrB with its conserved C-terminal cysteine
reaching the distal side of the siroheme. We propose a novel mechanism for the
process of sulfite reduction involving DsrAB, DsrC, and the DsrMKJOP membrane
complex (a membrane complex with putative disulfide/thiol reductase activity),
in which two of the six electrons for reduction of sulfite derive from the
membrane quinone pool. These results show that DsrC is involved in sulfite
reduction, which changes the mechanism of sulfate respiration. This has
important implications for models used to date ancient sulfur metabolism based
on sulfur isotope fractionations.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Structure of the DsrAB sulfite reductase bound to DsrC. A,
secondary structure representation of the α[2]β[2]γ[2]
assembly (DsrAB sulfite reductase bound to DsrC), with the
cofactors in ball-and-stick mode. DsrA (chains A and D) is
colored blue, DsrB (chains B and E) is magenta, and DsrC (chains
C and F) is green. The distance between the cofactors from one
αβγ unit is displayed on the right side. Color code is
yellow, carbon; red, oxygen; blue, nitrogen; brown, iron; and
green, sulfur. B, molecular surface of theα[2]β[2]γ[2]
assembly with oneαβγ unit in gray and the other colored
according to A. C, superposition of DsrA and DsrB. N-term, N
terminus; C-term, C terminus.
|
 |
Figure 3.
Substrate and DsrC-binding channels. A, molecular surface of
one αβγ unit showing the substrate channel, with a zoomed
view of the channel entrance, containing a randomly placed  ion
for scale; the distal site of the siroheme (in yellow) is
solvent-accessible. The color scheme is as in Fig. 1. B, surface
representation of DsrAB with DsrC displaced from its binding
position. The siroheme (in yellow) can be seen in the interior
of the cleft formed between DsrAB. C, secondary structure view
of one DsrABC unit with A. fulgidus DsrC (PDB code: 1SAU)
superposed. The zoomed image shows the extended C-terminal arm
of the D. vulgaris DsrC reaching the heme and the retracted arm
from A. fulgidus DsrC. The two conserved cysteines of each DsrC
are represented in stick mode, a dashed black line showing the
close contact between Cys-103 and Cys-114 in A. fulgidus DsrC.
Some water molecules at the interface are displayed as red
spheres. ion
for scale; the distal site of the siroheme (in yellow) is
solvent-accessible. The color scheme is as in Fig. 1. B, surface
representation of DsrAB with DsrC displaced from its binding
position. The siroheme (in yellow) can be seen in the interior
of the cleft formed between DsrAB. C, secondary structure view
of one DsrABC unit with A. fulgidus DsrC (PDB code: 1SAU)
superposed. The zoomed image shows the extended C-terminal arm
of the D. vulgaris DsrC reaching the heme and the retracted arm
from A. fulgidus DsrC. The two conserved cysteines of each DsrC
are represented in stick mode, a dashed black line showing the
close contact between Cys-103 and Cys-114 in A. fulgidus DsrC.
Some water molecules at the interface are displayed as red
spheres.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the ASBMB:
J Biol Chem
(2008,
283,
34141-34149)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.S.Habicht,
M.Miller,
R.P.Cox,
N.U.Frigaard,
M.Tonolla,
S.Peduzzi,
L.G.Falkenby,
and
J.S.Andersen
(2011).
Comparative proteomics and activity of a green sulfur bacterium through the water column of Lake Cadagno, Switzerland.
|
| |
Environ Microbiol,
13,
203-215.
|
 |
|
|
|
|
 |
M.Basen,
M.Krüger,
J.Milucka,
J.Kuever,
J.Kahnt,
O.Grundmann,
A.Meyerdierks,
F.Widdel,
and
S.Shima
(2011).
Bacterial enzymes for dissimilatory sulfate reduction in a marine microbial mat (Black Sea) mediating anaerobic oxidation of methane.
|
| |
Environ Microbiol,
13,
1370-1379.
|
 |
|
|
|
|
 |
F.Grimm,
N.Dobler,
and
C.Dahl
(2010).
Regulation of dsr genes encoding proteins responsible for the oxidation of stored sulfur in Allochromatium vinosum.
|
| |
Microbiology,
156,
764-773.
|
 |
|
|
|
|
 |
Y.C.Hsieh,
M.Y.Liu,
V.C.Wang,
Y.L.Chiang,
E.H.Liu,
W.G.Wu,
S.I.Chan,
and
C.J.Chen
(2010).
Structural insights into the enzyme catalysis from comparison of three forms of dissimilatory sulphite reductase from Desulfovibrio gigas.
|
| |
Mol Microbiol,
78,
1101-1116.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

