 |
PDBsum entry 2uuv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Alkyldihydroxyacetonephosphate synthase in p1
|
|
Structure:
|
 |
Alkyldihydroxyacetonephosphate synthase. Chain: a, b, c, d. Fragment: residues 9-587. Synonym: alkyl-dhap synthase, alkylglycerone-phosphate synthase. Engineered: yes
|
|
Source:
|
 |
Dictyostelium discoideum. Slime mold. Organism_taxid: 44689. Strain: kassx-3. Expressed in: escherichia coli. Expression_system_taxid: 469008. Expression_system_variant: plys. Other_details: academia DNA sequencing center in collaboration with dictyostelium cdna project
|
|
Resolution:
|
 |
|
1.99Å
|
R-factor:
|
0.228
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
A.Razeto,F.Mattiroli,E.Carpanelli,A.Aliverti,V.Pandini,A.Coda, A.Mattevi
|
Key ref:

|
 |
A.Razeto
et al.
(2007).
The crucial step in ether phospholipid biosynthesis: structural basis of a noncanonical reaction associated with a peroxisomal disorder.
Structure,
15,
683-692.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Mar-07
|
Release date:
|
26-Jun-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.2.5.1.26
- alkylglycerone-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a long chain fatty alcohol + a 1-acylglycerone 3-phosphate = a 1-O- alkylglycerone 3-phosphate + a long-chain fatty acid + H+
|
 |
 |
 |
 |
 |
long chain fatty alcohol
|
+
|
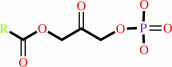 1-acylglycerone 3-phosphate
1-acylglycerone 3-phosphate
|
=
|
1-O- alkylglycerone 3-phosphate
|
+
|
 long-chain fatty acid
long-chain fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
15:683-692
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crucial step in ether phospholipid biosynthesis: structural basis of a noncanonical reaction associated with a peroxisomal disorder.
|
|
A.Razeto,
F.Mattiroli,
E.Carpanelli,
A.Aliverti,
V.Pandini,
A.Coda,
A.Mattevi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ether phospholipids are essential constituents of eukaryotic cell membranes.
Rhizomelic chondrodysplasia punctata type 3 is a severe peroxisomal disorder
caused by inborn deficiency of alkyldihydroxyacetonephosphate synthase (ADPS).
The enzyme carries out the most characteristic step in ether phospholipid
biosynthesis: formation of the ether bond. The crystal structure of ADPS from
Dictyostelium discoideum shows a fatty-alcohol molecule bound in a narrow
hydrophobic tunnel, specific for aliphatic chains of 16 carbons. Access to the
tunnel is controlled by a flexible loop and a gating helix at the
protein-membrane interface. Structural and mutagenesis investigations identify a
cluster of hydrophilic catalytic residues, including an essential tyrosine,
possibly involved in substrate proton abstraction, and the arginine that is
mutated in ADPS-deficient patients. We propose that ether bond formation might
be orchestrated through a covalent imine intermediate with the flavin,
accounting for the noncanonical employment of a flavin cofactor in a nonredox
reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Electrostatic Potentials of the ADPS Dimer Surface
Red, blue, and white show potentials at −8, +8, and 0
k[B]T e^−1, respectively. The basic amino acids forming the
membrane-binding region are labeled.
|
 |
Figure 7.
Figure 7. Structural Framework for ADPS Catalysis
(A) A
structural framework for the reaction cycle of ADPS. Step 1,
ADPS binds the acyl-DHAP substrate; the HHH loop becomes ordered
and the gating helix closes the tunnel. Step 2, the fatty-acid
product (R[1]COOH) is generated and released, whereas the DHAP
moiety remains trapped in the active site. Step 3, the fatty
alcohol (R[2]OH) binds in the tunnel. Step 4, the alkyl-DHAP
product is formed and released.
(B) Working hypothesis for
the reaction mechanism of ADPS.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2007,
15,
683-692)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Domínguez de María,
R.W.van Gemert,
A.J.Straathof,
and
U.Hanefeld
(2010).
Biosynthesis of ethers: unusual or common natural events?
|
| |
Nat Prod Rep,
27,
370-392.
|
 |
|
|
|
|
 |
F.Wang,
Z.Mei,
Y.Qi,
C.Yan,
S.Xiang,
Z.Zhou,
Q.Hu,
J.Wang,
and
Y.Shi
(2009).
Crystal structure of the MecA degradation tag.
|
| |
J Biol Chem,
284,
34376-34381.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Liefhebber,
B.W.Brandt,
R.Broer,
W.J.Spaan,
and
H.C.van Leeuwen
(2009).
Hepatitis C virus NS4B carboxy terminal domain is a membrane binding domain.
|
| |
Virol J,
6,
62.
|
 |
|
|
|
|
 |
F.Forneris,
and
A.Mattevi
(2008).
Enzymes without borders: mobilizing substrates, delivering products.
|
| |
Science,
321,
213-216.
|
 |
|
|
|
|
 |
D.E.Edmondson
(2007).
Plasmalogen assembly: a key flavoenzyme.
|
| |
Structure,
15,
639-641.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

