 |
PDBsum entry 2r2f

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2r2f
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
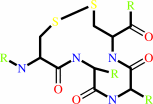 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
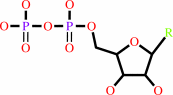 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
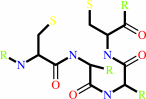 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
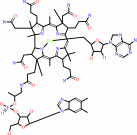 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:13359-13369
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of Salmonella typhimurium nrdF ribonucleotide reductase in its oxidized and reduced forms.
|
|
M.Eriksson,
A.Jordan,
H.Eklund.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The first class Ib ribonucleotide reductase R2 structure, from Salmonella
typhimurium, has been determined at 2.0 A resolution. The overall structure is
similar to the Escherichia coli class Ia enzyme despite only 23% sequence
identity. The most spectacular difference is the absence of the pleated sheet
and adjacent parts present in the E. coli R2 structure; the heart-shaped
structure loses its tip. From sequence comparisons, it appears that this feature
is shared with all other class Ib enzymes and, in this respect, is more like the
mammalian class Ia enzymes. Both the oxidized and reduced iron forms have been
investigated. In the ferric iron center, both iron ions are octahedrally
coordinated and bridged by one carboxylate and one oxide ion. The ferrous form
has lost the bridging oxide ion but is bridged by two carboxylates. Accompanying
the change in redox state, helix E changes its conformation from one covering
the metal center in the oxidized form to a more open reduced form. A narrow
channel is opened which may permit easier access of oxygen to the ferrous iron
site and to efficiently generate the tyrosyl radical.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Boal,
J.A.Cotruvo,
J.Stubbe,
and
A.C.Rosenzweig
(2010).
Structural basis for activation of class Ib ribonucleotide reductase.
|
| |
Science,
329,
1526-1530.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Johansson,
E.Torrents,
D.Lundin,
J.Sprenger,
M.Sahlin,
B.M.Sjöberg,
and
D.T.Logan
(2010).
High-resolution crystal structures of the flavoprotein NrdI in oxidized and reduced states--an unusual flavodoxin. Structural biology.
|
| |
FEBS J,
277,
4265-4277.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.A.Svistunenko,
and
G.A.Jones
(2009).
Tyrosyl radicals in proteins: a comparison of empirical and density functional calculated EPR parameters.
|
| |
Phys Chem Chem Phys,
11,
6600-6613.
|
 |
|
|
|
|
 |
J.A.Cotruvo,
and
J.Stubbe
(2008).
NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase.
|
| |
Proc Natl Acad Sci U S A,
105,
14383-14388.
|
 |
|
|
|
|
 |
W.Jiang,
D.Yun,
L.Saleh,
J.M.Bollinger,
and
C.Krebs
(2008).
Formation and function of the Manganese(IV)/Iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase.
|
| |
Biochemistry,
47,
13736-13744.
|
 |
|
|
|
|
 |
M.Galander,
M.Uppsten,
U.Uhlin,
and
F.Lendzian
(2006).
Orientation of the tyrosyl radical in Salmonella typhimurium class Ib ribonucleotide reductase determined by high field EPR of R2F single crystals.
|
| |
J Biol Chem,
281,
31743-31752.
|
 |
|
|
|
|
 |
M.H.Sazinsky,
P.W.Dunten,
M.S.McCormick,
A.DiDonato,
and
S.J.Lippard
(2006).
X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase.
|
| |
Biochemistry,
45,
15392-15404.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Nordlund,
and
P.Reichard
(2006).
Ribonucleotide reductases.
|
| |
Annu Rev Biochem,
75,
681-706.
|
 |
|
|
|
|
 |
E.Torrents,
M.Sahlin,
D.Biglino,
A.Gräslund,
and
B.M.Sjöberg
(2005).
Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical.
|
| |
Proc Natl Acad Sci U S A,
102,
17946-17951.
|
 |
|
|
|
|
 |
D.A.Svistunenko,
and
C.E.Cooper
(2004).
A new method of identifying the site of tyrosyl radicals in proteins.
|
| |
Biophys J,
87,
582-595.
|
 |
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
M.Kolberg,
A.K.Røhr,
C.H.Görbitz,
and
K.K.Andersson
(2004).
Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse.
|
| |
J Biol Chem,
279,
46794-46801.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Schwarzenbacher,
F.Stenner-Liewen,
H.Liewen,
H.Robinson,
H.Yuan,
E.Bossy-Wetzel,
J.C.Reed,
and
R.C.Liddington
(2004).
Structure of the Chlamydia protein CADD reveals a redox enzyme that modulates host cell apoptosis.
|
| |
J Biol Chem,
279,
29320-29324.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Schwarzenbacher,
F.Stenner-Liewen,
H.Liewen,
J.C.Reed,
and
R.C.Liddington
(2004).
Crystal structure of PqqC from Klebsiella pneumoniae at 2.1 A resolution.
|
| |
Proteins,
56,
401-403.
|
 |
|
|
|
|
 |
B.S.Cooperman
(2003).
Oligopeptide inhibition of class I ribonucleotide reductases.
|
| |
Biopolymers,
71,
117-131.
|
 |
|
|
|
|
 |
M.Uppsten,
M.Färnegårdh,
A.Jordan,
S.Ramaswamy,
and
U.Uhlin
(2003).
Expression and preliminary crystallographic studies of R1E, the large subunit of ribonucleotide reductase from Salmonella typhimurium.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1081-1083.
|
 |
|
|
|
|
 |
C.M.Summa,
A.Lombardi,
M.Lewis,
and
W.F.DeGrado
(1999).
Tertiary templates for the design of diiron proteins.
|
| |
Curr Opin Struct Biol,
9,
500-508.
|
 |
|
|
|
|
 |
M.Bolognesi,
C.Rosano,
R.Losso,
A.Borassi,
M.Rizzi,
J.B.Wittenberg,
A.Boffi,
and
P.Ascenzi
(1999).
Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
|
| |
Biophys J,
77,
1093-1099.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

