 |
PDBsum entry 2pb2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.11
- acetylornithine transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Ornithine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N2-acetyl-L-ornithine + 2-oxoglutarate = N-acetyl-L-glutamate 5-semialdehyde + L-glutamate
|
 |
 |
 |
 |
 |
N(2)-acetyl-L-ornithine
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 N-acetyl-L-glutamate 5-semialdehyde
N-acetyl-L-glutamate 5-semialdehyde
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.17
- succinyldiaminopimelate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-succinyl-(2S,6S)-2,6-diaminopimelate + 2-oxoglutarate = (S)-2- succinylamino-6-oxoheptanedioate + L-glutamate
|
 |
 |
 |
 |
 |
N-succinyl-(2S,6S)-2,6-diaminopimelate
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
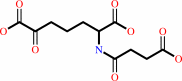 (S)-2- succinylamino-6-oxoheptanedioate
(S)-2- succinylamino-6-oxoheptanedioate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
70:429-441
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding.
|
|
V.Rajaram,
P.Ratna Prasuna,
H.S.Savithri,
M.R.Murthy.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acetylornithine aminotransferase (AcOAT) is one of the key enzymes involved in
arginine metabolism and catalyzes the conversion of N-acetylglutamate
semialdehyde to N-acetylornithine (AcOrn) in the presence of L-glutamate. It
belongs to the Type I subgroup II family of pyridoxal 5'-phosphate (PLP)
dependent enzymes. E. coli biosynthetic AcOAT (eAcOAT) also catalyzes the
conversion of N-succinyl-L-2-amino-6-oxopimelate to
N-succinyl-L,L-diaminopimelate, one of the steps in lysine biosynthesis. In view
of the critical role of AcOAT in lysine and arginine biosynthesis, structural
studies were initiated on the enzyme from S. typhimurium (sAcOAT). The K(m) and
k(cat)/K(m) values determined with the purified sAcOAT suggested that the enzyme
had much higher affinity for AcOrn than for ornithine (Orn) and was more
efficient than eAcOAT. sAcOAT was inhibited by gabaculine (Gcn) with an
inhibition constant (K(i)) of 7 microM and a second-order rate constant (k(2))
of 0.16 mM(-1) s(-1). sAcOAT, crystallized in the unliganded form and in the
presence of Gcn or L-glutamate, diffracted to a maximum resolution of 1.90 A and
contained a dimer in the asymmetric unit. The structure of unliganded sAcOAT
showed significant electron density for PLP in only one of the subunits (subunit
A). The asymmetry in PLP binding could be attributed to the ordering of the loop
L(alphak-) (betam) in only one subunit (subunit B; the loop from subunit B comes
close to the phosphate group of PLP in subunit A). Structural and spectral
studies of sAcOAT with Gcn suggested that the enzyme might have a low affinity
for PLP-Gcn complex. Comparison of sAcOAT with T. thermophilus AcOAT and human
ornithine aminotransferase suggested that the higher specificity of sAcOAT
towards AcOrn may not be due to specific changes in the active site residues but
could result from minor conformational changes in some of them. This is the
first structural report of AcOAT from a mesophilic organism and could serve as a
basis for drug design as the enzyme is important for bacterial cell wall
biosynthesis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. (a) Reaction catalyzed by AcOAT in arginine
biosynthesis. (b) Reaction catalyzed by AcOAT in lysine
biosynthesis. (c) Reaction of Gcn with PLP leading to the
formation of a Schiff base followed by proton abstraction and
aromatization to form m-carboxyphenylpyridoamine phosphate
(CPPP).
|
 |
Figure 9.
Figure 9. (a) Stereo view of electron density corresponding to
PLP in the active site of subunit A of sAcOAT-IA from the final
F[o]-F[c] map contoured at 3  .
Hydrogen-bonding interactions of PLP with the enzyme are shown
as dotted lines. (b) Stereo view of electron density
corresponding to PLP in the active site of subunit A of
sAcOAT-GCN in the final F[o]-F[c] map contoured at 3 .
Hydrogen-bonding interactions of PLP with the enzyme are shown
as dotted lines. (b) Stereo view of electron density
corresponding to PLP in the active site of subunit A of
sAcOAT-GCN in the final F[o]-F[c] map contoured at 3  .
PLP in sAcOAT-GCN is shown in light gray as a ball and stick
model and PLP from sAcOAT-IA is shown in dark gray. The small
tilt in orientation (7°) with respect to each other may be
observed. .
PLP in sAcOAT-GCN is shown in light gray as a ball and stick
model and PLP from sAcOAT-IA is shown in dark gray. The small
tilt in orientation (7°) with respect to each other may be
observed.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2008,
70,
429-441)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Stránská,
D.Kopecný,
M.Tylichová,
J.Snégaroff,
and
M.Sebela
(2008).
Ornithine delta-aminotransferase: An enzyme implicated in salt tolerance in higher plants.
|
| |
Plant Signal Behav,
3,
929-935.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

