 |
PDBsum entry 2jlc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.9
- 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic-acid
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isochorismate + 2-oxoglutarate + H+ = 5-enolpyruvoyl-6-hydroxy-2- succinyl-cyclohex-3-ene-1-carboxylate + CO2
|
 |
 |
 |
 |
 |
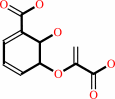 isochorismate
isochorismate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
5-enolpyruvoyl-6-hydroxy-2- succinyl-cyclohex-3-ene-1-carboxylate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
384:1353-1368
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Specificity and reactivity in menaquinone biosynthesis: the structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase).
|
|
A.Dawson,
P.K.Fyfe,
W.N.Hunter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The thiamine diphosphate (ThDP) and metal-ion-dependent enzyme
2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase, or
MenD, catalyze the Stetter-like conjugate addition of alpha-ketoglutarate with
isochorismate to release
2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate and carbon
dioxide. This reaction represents the first committed step for biosynthesis of
menaquinone, or vitamin K2, a key cofactor for electron transport in bacteria
and a metabolite for posttranslational modification of proteins in mammals. The
medium-resolution structure of MenD from Escherichia coli (EcMenD) in complex
with its cofactor and Mn2+ has been determined in two related hexagonal crystal
forms. The subunit displays the typical three-domain structure observed for
ThDP-dependent enzymes in which two of the domains bind and force the cofactor
into a configuration that supports formation of a reactive ylide. The structures
reveal a stable dimer-of-dimers association in agreement with gel filtration and
analytical ultracentrifugation studies and confirm the classification of MenD in
the pyruvate oxidase family of ThDP-dependent enzymes. The active site, created
by contributions from a pair of subunits, is highly basic with a pronounced
hydrophobic patch. These features, formed by highly conserved amino acids, match
well to the chemical properties of the substrates. A model of the covalent
intermediate formed after reaction with the first substrate alpha-ketoglutarate
and with the second substrate isochorismate positioned to accept nucleophilic
attack has been prepared. This, in addition to structural and sequence
comparisons with putative MenD orthologues, provides insight into the
specificity and reactivity of MenD and allows a two-stage reaction mechanism to
be proposed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Fig. 7. A stereoview of the model for substrates binding to
EcMenD. The post-decarboxylation covalent adduct of ThDP and
α-ketoglutarate (ThDP  asterisk
) is shown, and the reactive C atoms of this intermediate and
isochorismate are 1.6 Å apart. This separation is
indicated by a magenta broken line. The polypeptide main chain
is shown as a gray ribbon and atomic positions are colored as in
the previous figures except that the C atoms of the second
substrate, isochorismate, are yellow. A calculated electrostatic
surface potential (blue, positive; red, negative; white,
neutral) showing the active-site cleft. The figure was produced
using PyMOL^1 with Adaptive Poisson–Boltzmann Solver^39 with
electrostatic potential isocontours set at + 5 kT/e (blue) and
− 5 kT/e (red). asterisk
) is shown, and the reactive C atoms of this intermediate and
isochorismate are 1.6 Å apart. This separation is
indicated by a magenta broken line. The polypeptide main chain
is shown as a gray ribbon and atomic positions are colored as in
the previous figures except that the C atoms of the second
substrate, isochorismate, are yellow. A calculated electrostatic
surface potential (blue, positive; red, negative; white,
neutral) showing the active-site cleft. The figure was produced
using PyMOL^1 with Adaptive Poisson–Boltzmann Solver^39 with
electrostatic potential isocontours set at + 5 kT/e (blue) and
− 5 kT/e (red).
|
 |
Figure 8.
Fig. 8. A two-stage mechanism for catalysis by EcMenD. An
asterisk marks the isochorismate C2, which is attacked by the
carbanion intermediate.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
384,
1353-1368)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Dawson,
M.Chen,
P.K.Fyfe,
Z.Guo,
and
W.N.Hunter
(2010).
Structure and reactivity of Bacillus subtilis MenD catalyzing the first committed step in menaquinone biosynthesis.
|
| |
J Mol Biol,
401,
253-264.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.K.Fyfe,
A.Dawson,
M.T.Hutchison,
S.Cameron,
and
W.N.Hunter
(2010).
Structure of Staphylococcus aureus adenylosuccinate lyase (PurB) and assessment of its potential as a target for structure-based inhibitor discovery.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
881-888.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

