 |
PDBsum entry 2dfc
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
Structure of an orthorhombic form of xylanase ii from trichoderma reesei and analysis of thermal displacement.
|
 |
|
Authors
|
 |
N.Watanabe,
T.Akiba,
R.Kanai,
K.Harata.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 2006,
62,
784-792.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
Abstract
|
 |
|
An orthorhombic crystal of xylanase II from Trichoderma reesei was grown in the
presence of sodium iodide. Crystal structures at atomic resolution were
determined at 100 and 293 K. Protein molecules were aligned along a
crystallographic twofold screw axis, forming a helically extended polymer-like
chain mediated by an iodide ion. The iodide ion connected main-chain peptide
groups between two adjacent molecules by an N-H...I-...H-N hydrogen-bond bridge,
thus contributing to regulation of the molecular arrangement and suppression of
the rigid-body motion in the crystal with high diffraction quality. The
structure at 293 K showed considerable thermal motion in the loop regions
connecting the beta-strands that form the active-site cleft. TLS model analysis
of the thermal motion and a comparison between this structure and that at 100 K
suggest that the fluctuation of these loop regions is attributable to the
hinge-like movement of the beta-strands.
|
 |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 A stereoscopic view of hydrogen-bonding contacts of
iodide ions drawn with ORTEP-III (Burnett & Johnson,
1996[Burnett, M. N. & Johnson, C. K. (1996). ORTEP-III: Oak
Ridge Thermal Ellipsoid Plot Program for Crystal Structure
Illustrations. Report ORNL-6895, Oak Ridge National Laboratory,
Oak Ridge, TN, USA.]). Thermal ellipsoids are drawn with 30%
probability. The asterisks denote amino-acid residues of an
adjacent molecule.
|
 |
Figure 7.
Figure 7 Distance difference map (a) showing the movement of C^
 atoms
between the structures at 100 and 293 K. The distance difference
was calculated as the difference between the distances of C^ atoms
between the structures at 100 and 293 K. The distance difference
was calculated as the difference between the distances of C^
 atoms
as atoms
as 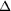 r
= r
=  C}_{\alpha}}]
(100 K) - C}_{\alpha}}]
(100 K) -  C}_{\alpha}}]
(293 K). Contours are drawn in the region from -0.4 to -1.8
Å with an interval of 0.2 Å. Loop regions
corresponding to the `fingertips' are denoted by T for the
`thumb' and A, B, C and D for the four `fingers'. Schematic
drawing of the energy landscape (b) shows the change of the
population of the atomic position of the `fingertip' regions
induced by cooling from 293 to 100 K. An average position is
denoted by x. C}_{\alpha}}]
(293 K). Contours are drawn in the region from -0.4 to -1.8
Å with an interval of 0.2 Å. Loop regions
corresponding to the `fingertips' are denoted by T for the
`thumb' and A, B, C and D for the four `fingers'. Schematic
drawing of the energy landscape (b) shows the change of the
population of the atomic position of the `fingertip' regions
induced by cooling from 293 to 100 K. An average position is
denoted by x.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
784-792)
copyright 2006.
|
 |
|

|
|
|
 |

