 |
PDBsum entry 2afx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human glutaminyl cyclase in complex with 1- benzylimidazole
|
|
Structure:
|
 |
Glutaminyl-peptide cyclotransferase. Chain: a, b. Fragment: residues 33-361. Synonym: qc, glutaminyl-tRNA cyclotransferase, glutaminyl cyclase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: qpct. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Hexamer (from
)
Hexamer (from
)
|
|
Resolution:
|
 |
|
1.64Å
|
R-factor:
|
0.177
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
K.F.Huang,Y.L.Liu,W.J.Cheng,T.P.Ko,A.H.J.Wang
|
Key ref:

|
 |
K.F.Huang
et al.
(2005).
Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation.
Proc Natl Acad Sci U S A,
102,
13117-13122.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Jul-05
|
Release date:
|
23-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16769
(QPCT_HUMAN) -
Glutaminyl-peptide cyclotransferase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
361 a.a.
323 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.2.5
- glutaminyl-peptide cyclotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-terminal L-glutaminyl-[peptide] = N-terminal 5-oxo-L-prolyl-[peptide] + NH4+
|
 |
 |
 |
 |
 |
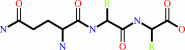 L-glutaminyl-peptide
L-glutaminyl-peptide
|
=
|
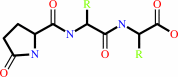 5-oxoprolyl-peptide
5-oxoprolyl-peptide
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
102:13117-13122
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation.
|
|
K.F.Huang,
Y.L.Liu,
W.J.Cheng,
T.P.Ko,
A.H.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
N-terminal pyroglutamate (pGlu) formation from its glutaminyl (or glutamyl)
precursor is required in the maturation of numerous bioactive peptides. The
aberrant formation of pGlu may be related to several pathological processes,
such as osteoporosis and amyloidotic diseases. This N-terminal cyclization
reaction, once thought to proceed spontaneously, is greatly facilitated by the
enzyme glutaminyl cyclase (QC). To probe this important but poorly understood
modification, we present here the structure of human QC in free form and bound
to a substrate and three imidazole-derived inhibitors. The structure reveals an
alpha/beta scaffold akin to that of two-zinc exopeptidases but with several
insertions and deletions, particularly in the active-site region. The relatively
closed active site displays alternate conformations due to the different indole
orientations of Trp-207, resulting in two substrate (glutamine t-butyl
ester)-binding modes. The single zinc ion in the active site is coordinated to
three conserved residues and one water molecule, which is replaced by an
imidazole nitrogen upon binding of the inhibitors. Together with structural and
kinetic analyses of several active-site-mutant enzymes, a catalysis mechanism of
the formation of protein N-terminal pGlu is proposed. Our results provide a
structural basis for the rational design of inhibitors against QC-associated
disorders.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Structure of human QC. (A) A ribbon diagram of the
overall structure of human QC. The central six  -strands
are colored orange. The -strands
are colored orange. The  -helices located on the
top, bottom, and edge are colored cyan, magenta, and yellow,
respectively. The zinc ion is shown as a yellow sphere. The
zinc-coordinated residues, Arg-54 (genetic mutation to Trp
residue occurred frequently in adult women with osteoporosis),
and a sulfate ion are depicted with a ball-and-stick model. The
coils and loops adjacent to the catalytic center are painted
green, whereas those distant from the active site are colored
gray. Gray dots represent the disordered region of residues
183-188. (B) A topology diagram of the human QC structure. The
color codes for secondary structural elements are identical to
those in A.(C) A stereoview of the human QC catalytic region.
The active-site residues in conf-A are shown and labeled.
Possible hydrogen and coordination bonds are represented with
dotted lines colored cyan and yellow, respectively. The green
dotted lines depict the possibly unusual hydrogen bonds between
D305 and E201 (3.06 Å) and between D305 and D248 (2.53 Å). -helices located on the
top, bottom, and edge are colored cyan, magenta, and yellow,
respectively. The zinc ion is shown as a yellow sphere. The
zinc-coordinated residues, Arg-54 (genetic mutation to Trp
residue occurred frequently in adult women with osteoporosis),
and a sulfate ion are depicted with a ball-and-stick model. The
coils and loops adjacent to the catalytic center are painted
green, whereas those distant from the active site are colored
gray. Gray dots represent the disordered region of residues
183-188. (B) A topology diagram of the human QC structure. The
color codes for secondary structural elements are identical to
those in A.(C) A stereoview of the human QC catalytic region.
The active-site residues in conf-A are shown and labeled.
Possible hydrogen and coordination bonds are represented with
dotted lines colored cyan and yellow, respectively. The green
dotted lines depict the possibly unusual hydrogen bonds between
D305 and E201 (3.06 Å) and between D305 and D248 (2.53 Å).
|
 |
Figure 4.
Fig. 4. Structures of human QC bound to imidazole-derived
inhibitors. (A) The zinc-binding environment of the free-form
human QC. The 2F[o] - F[c] electron density maps (contoured at
1.0  ) (gray) corresponding
to the water molecules inside the active-site pocket are shown.
Representations of the models, hydrogen bonds, and coordination
bonds are identical to those in Fig. 1C. (B-D) Structures of
human QC bound to 1-vinylimidazole (1.68-Å resolution),
1-benzylimidazole (1.64-Å resolution), and N- ) (gray) corresponding
to the water molecules inside the active-site pocket are shown.
Representations of the models, hydrogen bonds, and coordination
bonds are identical to those in Fig. 1C. (B-D) Structures of
human QC bound to 1-vinylimidazole (1.68-Å resolution),
1-benzylimidazole (1.64-Å resolution), and N-  -acetylhistamine
(1.56-Å resolution), respectively. The 2F[o] - F[c] maps
(contoured at 1.0 -acetylhistamine
(1.56-Å resolution), respectively. The 2F[o] - F[c] maps
(contoured at 1.0  ) (magenta) for the
inhibitors are overlaid with the final refined models. Distances
for enzyme-inhibitor interaction are indicated in Å. ) (magenta) for the
inhibitors are overlaid with the final refined models. Distances
for enzyme-inhibitor interaction are indicated in Å.
|
 |
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Oakley,
M.Coggan,
and
P.G.Board
(2010).
Identification and characterization of gamma-glutamylamine cyclotransferase, an enzyme responsible for gamma-glutamyl-epsilon-lysine catabolism.
|
| |
J Biol Chem,
285,
9642-9648.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.R.Carrillo,
C.Parthier,
N.Jänckel,
J.Grandke,
M.Stelter,
S.Schilling,
M.Boehme,
P.Neumann,
R.Wolf,
H.U.Demuth,
M.T.Stubbs,
and
J.U.Rahfeld
(2010).
Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus.
|
| |
Biol Chem,
391,
1419-1428.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Koike,
A.Kidera,
and
M.Ota
(2009).
Alteration of oligomeric state and domain architecture is essential for functional transformation between transferase and hydrolase with the same scaffold.
|
| |
Protein Sci,
18,
2060-2066.
|
 |
|
|
|
|
 |
M.Calvaresi,
M.Garavelli,
and
A.Bottoni
(2008).
Computational evidence for the catalytic mechanism of glutaminyl cyclase. A DFT investigation.
|
| |
Proteins,
73,
527-538.
|
 |
|
|
|
|
 |
S.Schilling,
C.Wasternack,
and
H.U.Demuth
(2008).
Glutaminyl cyclases from animals and plants: a case of functionally convergent protein evolution.
|
| |
Biol Chem,
389,
983-991.
|
 |
|
|
|
|
 |
J.L.Pereira,
E.F.Noronha,
R.N.Miller,
and
O.L.Franco
(2007).
Novel insights in the use of hydrolytic enzymes secreted by fungi with biotechnological potential.
|
| |
Lett Appl Microbiol,
44,
573-581.
|
 |
|
|
|
|
 |
K.Tessmar-Raible
(2007).
The evolution of neurosecretory centers in bilaterian forebrains: insights from protostomes.
|
| |
Semin Cell Dev Biol,
18,
492-501.
|
 |
|
|
|
|
 |
S.Schilling,
I.Stenzel,
A.von Bohlen,
M.Wermann,
K.Schulz,
H.U.Demuth,
and
C.Wasternack
(2007).
Isolation and characterization of the glutaminyl cyclases from Solanum tuberosum and Arabidopsis thaliana: implications for physiological functions.
|
| |
Biol Chem,
388,
145-153.
|
 |
|
|
|
|
 |
T.Guevara,
N.Mallorquí-Fernández,
R.García-Castellanos,
S.García-Piqué,
G.Ebert Petersen,
C.Lauritzen,
J.Pedersen,
J.Arnau,
F.X.Gomis-Rüth,
and
M.Solà
(2006).
Papaya glutamine cyclotransferase shows a singular five-fold beta-propeller architecture that suggests a novel reaction mechanism.
|
| |
Biol Chem,
387,
1479-1486.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

