 |
PDBsum entry 2a2j

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2a2j
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of a putative pyridoxine 5'-phosphate oxidase (rv2607) from mycobacterium tuberculosis
|
|
Structure:
|
 |
Pyridoxamine 5'-phosphate oxidase. Chain: a, b. Synonym: pnp/pmp oxidase, pnpox. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 1773. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.213
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
J.-D.Pedelacq,B.-S.Rho,C.-Y.Kim,G.S.Waldo,T.P.Lekin,B.W.Segelke, B.Rupp,L.-W.Hung,S.-I.Kim,T.C.Terwilliger,Mycobacterium Tuberculosis Structural Proteomics Project (Xmtb)
|
Key ref:

|
 |
J.D.Pédelacq
et al.
(2006).
Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins,
62,
563-569.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
22-Jun-05
|
Release date:
|
23-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WIJ1
(PDXH_MYCTU) -
Pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
224 a.a.
203 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.5
- pyridoxal 5'-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
pyridoxamine 5'-phosphate + O2 + H2O = pyridoxal 5'-phosphate + H2O2 + NH4+
|
|
2.
|
pyridoxine 5'-phosphate + O2 = pyridoxal 5'-phosphate + H2O2
|
|
 |
 |
 |
 |
 |
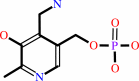 pyridoxamine 5'-phosphate
pyridoxamine 5'-phosphate
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
 pyridoxal 5'-phosphate
pyridoxal 5'-phosphate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
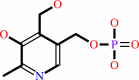 pyridoxine 5'-phosphate
pyridoxine 5'-phosphate
|
+
|
 O2
O2
|
=
|
 pyridoxal 5'-phosphate
pyridoxal 5'-phosphate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
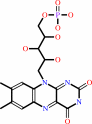 FMN
FMN
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
62:563-569
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
|
|
J.D.Pédelacq,
B.S.Rho,
C.Y.Kim,
G.S.Waldo,
T.P.Lekin,
B.W.Segelke,
B.Rupp,
L.W.Hung,
S.I.Kim,
T.C.Terwilliger.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structure of Rv2607, a putative pyridoxine 5'-phosphate
oxidase (PNPOx) from Mycobacterium tuberculosis, has been determined by X-ray
crystallography to 2.5 A resolution. Rv2607 has a core domain similar to known
PNPOx structures with a flavin mononucleotide (FMN) cofactor. Electron density
for two FMN at the dimer interface is weak despite the bright yellow color of
the protein solution and crystal. The shape and size of the putative binding
pocket is markedly different from that of members of the PNPOx family, which may
indicate some significant changes in the FMN binding mode of this protein
relative to members of the family.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Representation of the three-dimensional structure of
(A) Rv2607 and (B) the E. coli oxidase (PDB code: 1G79). (C)
Structure-based sequence alignment.  strands
are shown as arrows and strands
are shown as arrows and  helices
as coils. Sequence homologies are highlighted in red; sequence
identities are shown as white letters on a red background.
Residues important for FMN binding, PLP binding, and both, are
indicated using yellow, blue, and green triangles, respectively. helices
as coils. Sequence homologies are highlighted in red; sequence
identities are shown as white letters on a red background.
Residues important for FMN binding, PLP binding, and both, are
indicated using yellow, blue, and green triangles, respectively.
|
 |
Figure 4.
Figure 4. Stereo view of the active site. (A) Stereo diagrams
of 2Fo-Fc map at 1  level
(blue) and Fo-Fc map at 3 level
(blue) and Fo-Fc map at 3  level
(red). Only the residues within 5 Å of the FMN in Rv2607
are shown in ball-and-stick representation. (B) Superimposition
of Rv2607 (yellow) and the E. coli oxidase (pink) with FMN (PDB
code: 1G79). (C) Superimposition of Rv2607 (yellow) and the E.
coli oxidase (pink) in complex with PLP (PDB code: 1G79). level
(red). Only the residues within 5 Å of the FMN in Rv2607
are shown in ball-and-stick representation. (B) Superimposition
of Rv2607 (yellow) and the E. coli oxidase (pink) with FMN (PDB
code: 1G79). (C) Superimposition of Rv2607 (yellow) and the E.
coli oxidase (pink) in complex with PLP (PDB code: 1G79).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2006,
62,
563-569)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.N.Musayev,
M.L.Di Salvo,
M.A.Saavedra,
R.Contestabile,
M.S.Ghatge,
A.Haynes,
V.Schirch,
and
M.K.Safo
(2009).
Molecular basis of reduced pyridoxine 5'-phosphate oxidase catalytic activity in neonatal epileptic encephalopathy disorder.
|
| |
J Biol Chem,
284,
30949-30956.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.H.Huang,
R.J.Shi,
J.Y.Zhang,
Z.Wang,
and
L.Q.Huang
(2009).
Cloning and characterization of a pyridoxine 5'-phosphate oxidase from silkworm, Bombyx mori.
|
| |
Insect Mol Biol,
18,
365-371.
|
 |
|
|
|
|
 |
B.K.Biswal,
K.Au,
M.M.Cherney,
C.Garen,
and
M.N.James
(2006).
The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
735-742.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

