 |
PDBsum entry 1zlp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.7.1.1
- oxaloacetase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxaloacetate + H2O = oxalate + acetate + H+
|
 |
 |
 |
 |
 |
oxaloacetate
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
H2O
|
=
|
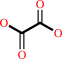 oxalate
oxalate
|
+
|
 acetate
acetate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.1.3.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
44:16377-16384
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the petal death protein from carnation flower.
|
|
A.Teplyakov,
S.Liu,
Z.Lu,
A.Howard,
D.Dunaway-Mariano,
O.Herzberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Expression of the PSR132 protein from Dianthus caryophyllus (carnation, clover
pink) is induced in response to ethylene production associated with petal
senescence, and thus the protein is named petal death protein (PDP). Recent work
has established that despite the annotation of PDP in sequence databases as
carboxyphosphoenolpyruvate mutase, the enzyme is actually a C-C bond cleaving
lyase exhibiting a broad substrate profile. The crystal structure of PDP has
been determined at 2.7 A resolution, revealing a dimer-of-dimers oligomeric
association. Consistent with sequence homology, the overall alpha/beta barrel
fold of PDP is the same as that of other isocitrate lyase/PEP mutase superfamily
members, including a swapped eighth helix within a dimer. Moreover, Mg(2+) binds
in the active site of PDP with a coordination pattern similar to that seen in
other superfamily members. A compound, covalently bound to the catalytic
residue, Cys144, was interpreted as a thiohemiacetal adduct resulting from the
reaction of glutaraldehyde used to cross-link the crystals. The Cys144-carrying
flexible loop that gates access to the active site is in the closed
conformation. Models of bound substrates and comparison with the closed
conformation of isocitrate lyase and 2-methylisocitrate lyase revealed the
structural basis for the broad substrate profile of PDP.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.C.Narayanan,
W.Niu,
Y.Han,
J.Zou,
P.S.Mariano,
D.Dunaway-Mariano,
and
O.Herzberg
(2008).
Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily.
|
| |
Biochemistry,
47,
167-182.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.J.Liao,
K.H.Chin,
C.H.Lin,
P.S.Tsai,
P.C.Lyu,
C.C.Young,
A.H.Wang,
and
S.H.Chou
(2008).
Crystal structure of DFA0005 complexed with alpha-ketoglutarate: a novel member of the ICL/PEPM superfamily from alkali-tolerant Deinococcus ficus.
|
| |
Proteins,
73,
362-371.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.J.Joosten,
Y.Han,
W.Niu,
J.Vervoort,
D.Dunaway-Mariano,
and
P.J.Schaap
(2008).
Identification of fungal oxaloacetate hydrolyase within the isocitrate lyase/PEP mutase enzyme superfamily using a sequence marker-based method.
|
| |
Proteins,
70,
157-166.
|
 |
|
|
|
|
 |
Y.Han,
H.J.Joosten,
W.Niu,
Z.Zhao,
P.S.Mariano,
M.McCalman,
J.van Kan,
P.J.Schaap,
and
D.Dunaway-Mariano
(2007).
Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi.
|
| |
J Biol Chem,
282,
9581-9590.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

