 |
PDBsum entry 1v2h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human pnp complexed with guanine
|
|
Structure:
|
 |
Purine nucleoside phosphorylase. Chain: e. Synonym: inosine phosphorylase, pnp. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pnp. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Trimer (from PDB file)
Trimer (from PDB file)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.218
|
R-free:
|
0.293
|
|
|
Authors:
|
 |
W.F.De Azevedo Jr.,F.Canduri,J.H.Pereira,D.M.Dos Santos,M.V.Bertacine Dias,R.G.Silva,M.S.Palma,L.A.Basso,D.S.Santos
|
|
Key ref:
|
 |
W.F.de Azevedo
et al.
(2003).
Crystal structure of human PNP complexed with guanine.
Biochem Biophys Res Commun,
312,
767-772.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Oct-03
|
Release date:
|
13-Jan-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00491
(PNPH_HUMAN) -
Purine nucleoside phosphorylase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
289 a.a.
288 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
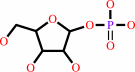 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
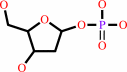 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochem Biophys Res Commun
312:767-772
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human PNP complexed with guanine.
|
|
W.F.de Azevedo,
F.Canduri,
D.M.dos Santos,
J.H.Pereira,
M.V.Bertacine Dias,
R.G.Silva,
M.A.Mendes,
L.A.Basso,
M.S.Palma,
D.S.Santos.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine nucleoside phosphorylase (PNP) catalyzes the phosphorolysis of the
N-ribosidic bonds of purine nucleosides and deoxynucleosides. PNP is a target
for inhibitor development aiming at T-cell immune response modulation and has
been submitted to extensive structure-based drug design. More recently, the 3-D
structure of human PNP has been refined to 2.3A resolution, which allowed a
redefinition of the residues involved in the substrate-binding sites and
provided a more reliable model for structure-based design of inhibitors. This
work reports crystallographic study of the complex of Human PNP:guanine
(HsPNP:Gua) solved at 2.7A resolution using synchrotron radiation. Analysis of
the structural differences among the HsPNP:Gua complex, PNP apoenzyme, and
HsPNP:immucillin-H provides explanation for inhibitor binding, refines the
purine-binding site, and can be used for future inhibitor design.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.B.Zanchi,
R.A.Caceres,
R.G.Stabeli,
and
W.F.de Azevedo
(2010).
Molecular dynamics studies of a hexameric purine nucleoside phosphorylase.
|
| |
J Mol Model,
16,
543-550.
|
 |
|
|
|
|
 |
I.Pauli,
L.F.Timmers,
R.A.Caceres,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2009).
Molecular modeling and dynamics studies of purine nucleoside phosphorylase from Bacteroides fragilis.
|
| |
J Mol Model,
15,
913-922.
|
 |
|
|
|
|
 |
F.Canduri,
R.G.Silva,
D.M.dos Santos,
M.S.Palma,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2005).
Structure of human PNP complexed with ligands.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
856-862.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

