 |
PDBsum entry 1v45

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human pnp complexed with 3-deoxyguanosine
|
|
Structure:
|
 |
Purine nucleoside phosphorylase. Chain: e. Synonym: inosine phosphorylase, pnp. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Trimer (from PDB file)
Trimer (from PDB file)
|
|
Resolution:
|
 |
|
2.86Å
|
R-factor:
|
0.180
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
F.Canduri,D.M.Dos Santos,R.G.Silva,M.A.Mendes,M.S.Palma,W.F.De Azevedo Jr.,L.A.Basso,D.S.Santos
|
Key ref:

|
 |
F.Canduri
et al.
(2005).
Structure of human PNP complexed with ligands.
Acta Crystallogr D Biol Crystallogr,
61,
856-862.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Nov-03
|
Release date:
|
14-Dec-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00491
(PNPH_HUMAN) -
Purine nucleoside phosphorylase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
289 a.a.
288 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
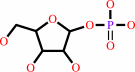 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
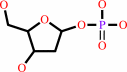 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
61:856-862
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of human PNP complexed with ligands.
|
|
F.Canduri,
R.G.Silva,
D.M.dos Santos,
M.S.Palma,
L.A.Basso,
D.S.Santos,
W.F.de Azevedo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine nucleoside phosphorylase (PNP) is a key enzyme in the purine-salvage
pathway, which allows cells to utilize preformed bases and nucleosides in order
to synthesize nucleotides. PNP is specific for purine nucleosides in the
beta-configuration and exhibits a strong preference for purines containing a
6-keto group and ribosyl-containing nucleosides relative to the corresponding
analogues. PNP was crystallized in complex with ligands and data collection was
performed using synchrotron radiation. This work reports the structure of human
PNP in complex with guanosine (at 2.80 A resolution), 3'-deoxyguanosine (at 2.86
A resolution) and 8-azaguanine (at 2.85 A resolution). These structures were
compared with the PNP-guanine, PNP-inosine and PNP-immucillin-H complexes solved
previously.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Superposition of PNP apoenzyme with the PNP-guanine
complex, showing the gate movement, which involves a helical
transformation of residues 257-265 in the transition apoenzyme
complex. Movements of the residues Tyr249, His257 and Lys265 and
the position of ligand and sulfates can be observed. The figure
was generated with the program MOLMOL (Koradi et al.,
1996[Koradi, R., Billeter, M. & Wuthrich, K. (1996). J. Mol.
Graph. 14, 51-55.]).
|
 |
Figure 4.
Figure 4 Binding site of human PNP. (a) HsPNP-guanosine, (b)
HsPNP-inosine, (c) HsPNP-immucillin-H, (d)
HsPNP-3'-deoxyguanosine, (e) HsPNP-8-azaguanine, (f)
HsPNP-guanine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2005,
61,
856-862)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.B.Zanchi,
R.A.Caceres,
R.G.Stabeli,
and
W.F.de Azevedo
(2010).
Molecular dynamics studies of a hexameric purine nucleoside phosphorylase.
|
| |
J Mol Model,
16,
543-550.
|
 |
|
|
|
|
 |
M.L.Bellows,
and
C.A.Floudas
(2010).
Computational methods for de novo protein design and its applications to the human immunodeficiency virus 1, purine nucleoside phosphorylase, ubiquitin specific protease 7, and histone demethylases.
|
| |
Curr Drug Targets,
11,
264-278.
|
 |
|
|
|
|
 |
I.Pauli,
L.F.Timmers,
R.A.Caceres,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2009).
Molecular modeling and dynamics studies of purine nucleoside phosphorylase from Bacteroides fragilis.
|
| |
J Mol Model,
15,
913-922.
|
 |
|
|
|
|
 |
S.Afshar,
M.R.Sawaya,
and
S.L.Morrison
(2009).
Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2'-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine.
|
| |
Protein Sci,
18,
1107-1114.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Afshar,
T.Asai,
and
S.L.Morrison
(2009).
Humanized ADEPT comprised of an engineered human purine nucleoside phosphorylase and a tumor targeting peptide for treatment of cancer.
|
| |
Mol Cancer Ther,
8,
185-193.
|
 |
|
|
|
|
 |
S.Afshar,
T.Olafsen,
A.M.Wu,
and
S.L.Morrison
(2009).
Characterization of an engineered human purine nucleoside phosphorylase fused to an anti-her2/neu single chain Fv for use in ADEPT.
|
| |
J Exp Clin Cancer Res,
28,
147.
|
 |
|
|
|
|
 |
J.E.Hyde
(2007).
Targeting purine and pyrimidine metabolism in human apicomplexan parasites.
|
| |
Curr Drug Targets,
8,
31-47.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

