 |
PDBsum entry 1us0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1us0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Human aldose reductase in complex with NADP+ and the inhibitor idd594 at 0.66 angstrom
|
|
Structure:
|
 |
Aldose reductase. Chain: a. Synonym: aldehyde reductase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
0.66Å
|
R-factor:
|
0.094
|
R-free:
|
0.103
|
|
|
Authors:
|
 |
E.I.Howard,R.Sanishvili,R.E.Cachau,A.Mitschler,B.Chevrier,P.Barth, V.Lamour,M.Van Zandt,E.Sibley,C.Bon,D.Moras,T.R.Schneider, A.Joachimiak,A.Podjarny
|
Key ref:

|
 |
E.I.Howard
et al.
(2004).
Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A.
Proteins,
55,
792-804.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Nov-03
|
Release date:
|
07-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15121
(ALDR_HUMAN) -
Aldo-keto reductase family 1 member B1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
316 a.a.
314 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
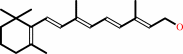 all-trans-retinol
all-trans-retinol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
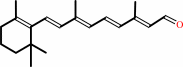 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
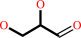 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
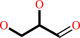 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
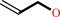 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
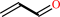 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
55:792-804
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A.
|
|
E.I.Howard,
R.Sanishvili,
R.E.Cachau,
A.Mitschler,
B.Chevrier,
P.Barth,
V.Lamour,
M.Van Zandt,
E.Sibley,
C.Bon,
D.Moras,
T.R.Schneider,
A.Joachimiak,
A.Podjarny.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The first subatomic resolution structure of a 36 kDa protein [aldose reductase
is presented. AR was cocrystallized at pH 5.0 with its cofactor NADP+ and
inhibitor IDD 594, a therapeutic candidate for the treatment of diabetic
complications. X-ray diffraction data were collected up to 0.62 A resolution and
treated up to 0.66 A resolution. Anisotropic refinement followed by a blocked
matrix inversion produced low standard deviations (<0.005 A). The model was
very well ordered overall (CA atoms' mean B factor is 5.5 A2). The model and the
electron-density maps revealed fine features, such as H-atoms, bond densities,
and significant deviations from standard stereochemistry. Other features, such
as networks of hydrogen bonds (H bonds), a large number of multiple
conformations, and solvent structure were also better defined. Most of the atoms
in the active site region were extremely well ordered (mean B approximately 3
A2), leading to the identification of the protonation states of the residues
involved in catalysis. The electrostatic interactions of the inhibitor's charged
carboxylate head with the catalytic residues and the charged coenzyme NADP+
explained the inhibitor's noncompetitive character. Furthermore, a short contact
involving the IDD 594 bromine atom explained the selectivity profile of the
inhibitor, important feature to avoid toxic effects. The presented structure and
the details revealed are instrumental for better understanding of the inhibition
mechanism of AR by IDD 594, and hence, for the rational drug design of future
inhibitors. This work demonstrates the capabilities of subatomic resolution
experiments and stimulates further developments of methods allowing the use of
the full potential of these experiments.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Contacts of IDD 594 with AR and NADP^+. (a) 3D view
of inhibitor contacts. View of the inhibitor-binding site down
the barrel axis with the semitransparent representation of AR
surface. The active site cleft (marked A), is occupied by the
inhibitor (stick only; color code: C = gray, N = blue, O = red,
F = pink, S = orange, Br = green) and surrounded by the
catalytic residues His 110, Tyr 48, and the coenzyme NADP^+. The
residues within 3.9 Å and NADP^+ are shown (balls and
sticks). The specificity pocket between Leu 300, Phe 122, and
Trp 111 (marked S) is occupied by the brominated aromatic ring
of the inhibitor. (b) Scheme of IDD 594 and its contacts. Red
dashed lines show contacts between 3.5 and 3.0 Å, and blue
dashed lines show contacts <3.0 Å. The exact values are
given in Table IV.
|
 |
Figure 5.
Figure 5. Model of residues Cys 44 and Ala 45 superimposed with
 A-weighted
Fo-Fc map with omitted hydrogen atoms (a), contoured magenta at
at 0.25 e/Å^3 (2.5 A-weighted
Fo-Fc map with omitted hydrogen atoms (a), contoured magenta at
at 0.25 e/Å^3 (2.5  )
showing the position of the hydrogen atoms and the bond
densities. Model of residues Ser 76 and Lys 77(b), viewed down
the C )
showing the position of the hydrogen atoms and the bond
densities. Model of residues Ser 76 and Lys 77(b), viewed down
the C  N
peptide bond showing the N
peptide bond showing the  angle
of -167.3° stabilized by an H-bond. angle
of -167.3° stabilized by an H-bond.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2004,
55,
792-804)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Sunitha,
H.C.Devarajegowda,
W.F.Al-Eryani,
Y.R.Prasad,
and
A.U.Kumar
(2012).
(2E)-1-(5-Bromo-thio-phen-2-yl)-3-(2,3,4-trimeth-oxy-phen-yl)prop-2-en-1-one.
|
| |
Acta Crystallogr Sect E Struct Rep Online,
68,
o61.
|
 |
|
|
|
|
 |
A.M.Katsori,
M.Chatzopoulou,
K.Dimas,
C.Kontogiorgis,
A.Patsilinakos,
T.Trangas,
and
D.Hadjipavlou-Litina
(2011).
Curcumin analogues as possible anti-proliferative & anti-inflammatory agents.
|
| |
Eur J Med Chem,
46,
2722-2735.
|
 |
|
|
|
|
 |
E.Parisini,
P.Metrangolo,
T.Pilati,
G.Resnati,
and
G.Terraneo
(2011).
Halogen bonding in halocarbon-protein complexes: a structural survey.
|
| |
Chem Soc Rev,
40,
2267-2278.
|
 |
|
|
|
|
 |
G.M.Alushin,
D.Jane,
and
M.L.Mayer
(2011).
Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
|
| |
Neuropharmacology,
60,
126-134.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Higashiura,
T.Kurakane,
M.Matsuda,
M.Suzuki,
K.Inaka,
M.Sato,
T.Kobayashi,
T.Tanaka,
H.Tanaka,
K.Fujiwara,
and
A.Nakagawa
(2010).
High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
698-708.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Gabison,
M.Chiadmi,
M.El Hajji,
B.Castro,
N.Colloc'h,
and
T.Prangé
(2010).
Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
714-724.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.K.Jiang
(2010).
Molecular docking and 3D-QSAR studies on beta-phenylalanine derivatives as dipeptidyl peptidase IV inhibitors.
|
| |
J Mol Model,
16,
1239-1249.
|
 |
|
|
|
|
 |
Y.Lu,
Y.Wang,
and
W.Zhu
(2010).
Nonbonding interactions of organic halogens in biological systems: implications for drug discovery and biomolecular design.
|
| |
Phys Chem Chem Phys,
12,
4543-4551.
|
 |
|
|
|
|
 |
Z.Dauter,
M.Jaskolski,
and
A.Wlodawer
(2010).
Impact of synchrotron radiation on macromolecular crystallography: a personal view.
|
| |
J Synchrotron Radiat,
17,
433-444.
|
 |
|
|
|
|
 |
A.S.Gardberg,
M.P.Blakeley,
and
D.A.Myles
(2009).
A preliminary neutron crystallographic study of proteinase K at pD 6.5.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
184-187.
|
 |
|
|
|
|
 |
A.Urzhumtsev,
P.V.Afonine,
and
P.D.Adams
(2009).
On the use of logarithmic scales for analysis of diffraction data.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1283-1291.
|
 |
|
|
|
|
 |
E.Oksanen,
M.P.Blakeley,
F.Bonneté,
M.T.Dauvergne,
F.Dauvergne,
and
M.Budayova-Spano
(2009).
Large crystal growth by thermal control allows combined X-ray and neutron crystallographic studies to elucidate the protonation states in Aspergillus flavus urate oxidase.
|
| |
J R Soc Interface,
6,
S599-S610.
|
 |
|
|
|
|
 |
M.Eisenmann,
H.Steuber,
M.Zentgraf,
M.Altenkämper,
R.Ortmann,
J.Perruchon,
G.Klebe,
and
M.Schlitzer
(2009).
Structure-based optimization of aldose reductase inhibitors originating from virtual screening.
|
| |
ChemMedChem,
4,
809-819.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Schnieders,
T.D.Fenn,
V.S.Pande,
and
A.T.Brunger
(2009).
Polarizable atomic multipole X-ray refinement: application to peptide crystals.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
952-965.
|
 |
|
|
|
|
 |
V.B.Chen,
I.W.Davis,
and
D.C.Richardson
(2009).
KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program.
|
| |
Protein Sci,
18,
2403-2409.
|
 |
|
|
|
|
 |
B.Guillot,
C.Jelsch,
A.Podjarny,
and
C.Lecomte
(2008).
Charge-density analysis of a protein structure at subatomic resolution: the human aldose reductase case.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
567-588.
|
 |
|
|
|
|
 |
E.Nishibori,
T.Nakamura,
M.Arimoto,
S.Aoyagi,
H.Ago,
M.Miyano,
T.Ebisuzaki,
and
M.Sakata
(2008).
Application of maximum-entropy maps in the accurate refinement of a putative acylphosphatase using 1.3 A X-ray diffraction data.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
237-247.
|
 |
|
|
|
|
 |
M.P.Blakeley,
F.Ruiz,
R.Cachau,
I.Hazemann,
F.Meilleur,
A.Mitschler,
S.Ginell,
P.Afonine,
O.N.Ventura,
A.Cousido-Siah,
M.Haertlein,
A.Joachimiak,
D.Myles,
and
A.Podjarny
(2008).
Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
|
| |
Proc Natl Acad Sci U S A,
105,
1844-1848.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Niimura,
and
R.Bau
(2008).
Neutron protein crystallography: beyond the folding structure of biological macromolecules.
|
| |
Acta Crystallogr A,
64,
12-22.
|
 |
|
|
|
|
 |
A.A.Rashin,
and
A.H.Rashin
(2007).
Surface hydrophobic groups, stability, and flip-flopping in lattice proteins.
|
| |
Proteins,
66,
321-341.
|
 |
|
|
|
|
 |
A.Volkov,
M.Messerschmidt,
and
P.Coppens
(2007).
Improving the scattering-factor formalism in protein refinement: application of the University at Buffalo Aspherical-Atom Databank to polypeptide structures.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
160-170.
|
 |
|
|
|
|
 |
D.Rakowitz,
G.Piccolruaz,
C.Pirker,
and
B.Matuszczak
(2007).
Novel aldose reductase inhibitors derived from 6-[[(diphenylmethylene)amino]oxy]hexanoic acid.
|
| |
Arch Pharm (Weinheim),
340,
202-208.
|
 |
|
|
|
|
 |
H.U.Ahmed,
M.P.Blakeley,
M.Cianci,
D.W.Cruickshank,
J.A.Hubbard,
and
J.R.Helliwell
(2007).
The determination of protonation states in proteins.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
906-922.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wang,
M.Dauter,
R.Alkire,
A.Joachimiak,
and
Z.Dauter
(2007).
Triclinic lysozyme at 0.65 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
1254-1268.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Biadene,
I.Hazemann,
A.Cousido,
S.Ginell,
A.Joachimiak,
G.M.Sheldrick,
A.Podjarny,
and
T.R.Schneider
(2007).
The atomic resolution structure of human aldose reductase reveals that rearrangement of a bound ligand allows the opening of the safety-belt loop.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
665-672.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Jaskolski,
M.Gilski,
Z.Dauter,
and
A.Wlodawer
(2007).
Stereochemical restraints revisited: how accurate are refinement targets and how much should protein structures be allowed to deviate from them?
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
611-620.
|
 |
|
|
|
|
 |
D.Rakowitz,
A.Gmeiner,
and
B.Matuszczak
(2006).
Discovery of novel aldose reductase inhibitors characterized by an alkoxy-substituted phenylacetic acid core.
|
| |
Arch Pharm (Weinheim),
339,
559-563.
|
 |
|
|
|
|
 |
D.Rakowitz,
H.Angerer,
and
B.Matuszczak
(2006).
Synthesis and aldose reductase inhibitory activities of novel O-substituted hydroxyphenylacetic acid derivatives.
|
| |
Arch Pharm (Weinheim),
339,
547-558.
|
 |
|
|
|
|
 |
F.Meilleur,
D.A.Myles,
and
M.P.Blakeley
(2006).
Neutron Laue macromolecular crystallography.
|
| |
Eur Biophys J,
35,
611-620.
|
 |
|
|
|
|
 |
G.Rosenbaum,
R.W.Alkire,
G.Evans,
F.J.Rotella,
K.Lazarski,
R.G.Zhang,
S.L.Ginell,
N.Duke,
I.Naday,
J.Lazarz,
M.J.Molitsky,
L.Keefe,
J.Gonczy,
L.Rock,
R.Sanishvili,
M.A.Walsh,
E.Westbrook,
and
A.Joachimiak
(2006).
The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results.
|
| |
J Synchrotron Radiat,
13,
30-45.
|
 |
|
|
|
|
 |
J.Hakanpää,
M.Linder,
A.Popov,
A.Schmidt,
and
J.Rouvinen
(2006).
Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
356-367.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.M.Brownlee,
E.Carlson,
A.C.Milne,
E.Pape,
and
D.H.Harrison
(2006).
Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
|
| |
Bioorg Chem,
34,
424-444.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.P.Blakeley,
A.Mitschler,
I.Hazemann,
F.Meilleur,
D.A.Myles,
and
A.Podjarny
(2006).
Comparison of hydrogen determination with X-ray and neutron crystallography in a human aldose reductase-inhibitor complex.
|
| |
Eur Biophys J,
35,
577-583.
|
 |
|
|
|
|
 |
T.Petrova,
S.Ginell,
A.Mitschler,
I.Hazemann,
T.Schneider,
A.Cousido,
V.Y.Lunin,
A.Joachimiak,
and
A.Podjarny
(2006).
Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1535-1544.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.A.Rashin,
and
A.H.Rashin
(2005).
Lattice models, packing density, and Boltzmann-like distribution of cavities in proteins.
|
| |
Proteins,
58,
547-559.
|
 |
|
|
|
|
 |
D.Rakowitz,
P.Muigg,
N.Schröder,
and
B.Matuszczak
(2005).
On the synthesis of bioisosters of o-benzothiazolyloxybenzoic acids and evaluation as aldose reductase inhibitors.
|
| |
Arch Pharm (Weinheim),
338,
419-426.
|
 |
|
|
|
|
 |
H.Bönisch,
C.L.Schmidt,
P.Bianco,
and
R.Ladenstein
(2005).
Ultrahigh-resolution study on Pyrococcus abyssi rubredoxin. I. 0.69 A X-ray structure of mutant W4L/R5S.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
990.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Hazemann,
M.T.Dauvergne,
M.P.Blakeley,
F.Meilleur,
M.Haertlein,
A.Van Dorsselaer,
A.Mitschler,
D.A.Myles,
and
A.Podjarny
(2005).
High-resolution neutron protein crystallography with radically small crystal volumes: application of perdeuteration to human aldose reductase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1413-1417.
|
 |
|
|
|
|
 |
M.P.Blakeley,
A.J.Kalb,
J.R.Helliwell,
and
D.A.Myles
(2004).
The 15-K neutron structure of saccharide-free concanavalin A.
|
| |
Proc Natl Acad Sci U S A,
101,
16405-16410.
|
 |
|
|
|
|
 |
P.Auffinger,
F.A.Hays,
E.Westhof,
and
P.S.Ho
(2004).
Halogen bonds in biological molecules.
|
| |
Proc Natl Acad Sci U S A,
101,
16789-16794.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

