 |
PDBsum entry 1tt0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1tt0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.10
- pyranose oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + O2 = 2-dehydro-D-glucose + H2O2
|
 |
 |
 |
 |
 |
D-glucose
Bound ligand (Het Group name = )
matches with 41.18% similarity
|
+
|
 O2
O2
|
=
|
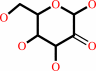 2-dehydro-D-glucose
2-dehydro-D-glucose
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
341:781-796
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase.
|
|
B.M.Hallberg,
C.Leitner,
D.Haltrich,
C.Divne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Pyranose 2-oxidase (P2Ox) is a 270 kDa homotetramer localized preferentially in
the hyphal periplasmic space of lignocellulolytic fungi and has a proposed role
in lignocellulose degradation to produce the essential co-substrate, hydrogen
peroxide, for lignin peroxidases. P2Ox oxidizes D-glucose and other
aldopyranoses regioselectively at C2 to the corresponding 2-keto sugars;
however, for some substrates, the enzyme also displays specificity for oxidation
at C3. The crystal structure of P2Ox from Trametes multicolor has been
determined using single anomalous dispersion with mercury as anomalous
scatterer. The model was refined at 1.8A resolution to R and Rfree values of
0.134 and 0.171, respectively. The overall fold of the P2Ox subunit resembles
that of members of the glucose-methanol-choline family of long-chain
oxidoreductases, featuring a flavin-binding Rossmann domain of class alpha/beta
and a substrate-binding subdomain with a six-stranded central beta sheet and
three alpha helices. The homotetramer buries a large internal cavity of roughly
15,000 A3, from which the four active sites are accessible. Four solvent
channels lead from the surface into the cavity through which substrate must
enter before accessing the active site. The present structure shows an acetate
molecule bound in the active site with the carboxylate group positioned
immediately below the flavin N5 atom, and with one carboxylate oxygen atom
interacting with the catalytic residues His548 and Asn593. The entrance to the
active site is blocked by a loop (residues 452 to 461) with excellent electron
density but elevated temperature factors. We predict that this loop is dynamic
and opens to allow substrate entry and exit. In silico docking of D-glucose in
the P2Ox active site shows that with the active-site loop in the closed
conformation, monosaccharides cannot be accommodated; however, after removing
the loop from the model, a tentative set of protein-substrate interactions for
beta-D-glucose have been outlined.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. The active site in P2Ox. A, The binding of
acetate in the P2Ox active site. Key residues that make up the
active site are shown. The active-site loop (residues 452-456)
that blocks the catalytic site from substrate access is
highlighted in purple. Atom-coloring scheme: carbon, beige
(protein), yellow (FAD), green (acetate); nitrogen, blue;
oxygen, red. For clarity of the picture, water molecules were
not included. B, Structural superposition of the oxidative site
in P2Ox (violet) and DH[cdh][41.] (green). The least-squares
comparison was made to optimize the overall superposition of the
active sites. The FAD co-factor molecules, the catalytic His-Asn
pairs (P2Ox residue numbering), and the ligands (P2Ox, acetate;
DH[cdh], cellobionolactam) are included, as well as Phe454 in
P2Ox, which assumes the approximate position of the non-reducing
end glucosyl moiety in the DH[cdh]-cellobionolactam complex.
|
 |
Figure 8.
Figure 8. Comparison of the active site in P2Ox with
related GMC enzymes. The FAD co-factor and the catalytic
residues are shown as stick models for P2Ox (violet), DH[cdh]
(green), ChOx (yellow) and GOx (orange). The active sites were
superimposed with respect to the flavin N5 atom. Residue
numbering for the catalytic His-Asn is that for P2Ox (for
details, see the text). The flavin ring in DH[cdh] is modified
by hydroxylation at C6.[27.] FAD-HNL was not included in the
comparison, since this enzyme is not an oxidoreductase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
341,
781-796)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Salaheddin,
Y.Takakura,
M.Tsunashima,
B.Stranzinger,
O.Spadiut,
M.Yamabhai,
C.K.Peterbauer,
and
D.Haltrich
(2010).
Characterisation of recombinant pyranose oxidase from the cultivated mycorrhizal basidiomycete Lyophyllum shimeji (hon-shimeji).
|
| |
Microb Cell Fact,
9,
57.
|
 |
|
|
|
|
 |
O.Spadiut,
T.C.Tan,
I.Pisanelli,
D.Haltrich,
and
C.Divne
(2010).
Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
|
| |
FEBS J,
277,
2892-2909.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Orville,
G.T.Lountos,
S.Finnegan,
G.Gadda,
and
R.Prabhakar
(2009).
Crystallographic, spectroscopic, and computational analysis of a flavin C4a-oxygen adduct in choline oxidase.
|
| |
Biochemistry,
48,
720-728.
|
 |
|
|
|
|
 |
I.Dreveny,
A.S.Andryushkova,
A.Glieder,
K.Gruber,
and
C.Kratky
(2009).
Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
|
| |
Biochemistry,
48,
3370-3377.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
C.Leitner,
C.Salaheddin,
B.Varga,
B.G.Vertessy,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design.
|
| |
FEBS J,
276,
776-792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
K.Radakovits,
I.Pisanelli,
C.Salaheddin,
M.Yamabhai,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
A thermostable triple mutant of pyranose 2-oxidase from Trametes multicolor with improved properties for biotechnological applications.
|
| |
Biotechnol J,
4,
525-534.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
K.Ida,
M.Kurabayashi,
M.Suguro,
Y.Hiruma,
T.Hikima,
M.Yamomoto,
and
H.Suzuki
(2008).
Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
|
| |
J Biol Chem,
283,
16584-16590.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Kujawa,
J.Volc,
P.Halada,
P.Sedmera,
C.Divne,
C.Sygmund,
C.Leitner,
C.Peterbauer,
and
D.Haltrich
(2007).
Properties of pyranose dehydrogenase purified from the litter-degrading fungus Agaricus xanthoderma.
|
| |
FEBS J,
274,
879-894.
|
 |
|
|
|
|
 |
M.Kujawa,
H.Ebner,
C.Leitner,
B.M.Hallberg,
M.Prongjit,
J.Sucharitakul,
R.Ludwig,
U.Rudsander,
C.Peterbauer,
P.Chaiyen,
D.Haltrich,
and
C.Divne
(2006).
Structural basis for substrate binding and regioselective oxidation of monosaccharides at C3 by pyranose 2-oxidase.
|
| |
J Biol Chem,
281,
35104-35115.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.H.Huang,
W.L.Lai,
M.H.Lee,
C.J.Chen,
A.Vasella,
Y.C.Tsai,
and
S.H.Liaw
(2005).
Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD.
|
| |
J Biol Chem,
280,
38831-38838.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.H.Lee,
W.L.Lai,
S.F.Lin,
C.S.Hsu,
S.H.Liaw,
and
Y.C.Tsai
(2005).
Structural characterization of glucooligosaccharide oxidase from Acremonium strictum.
|
| |
Appl Environ Microbiol,
71,
8881-8887.
|
 |
|
|
|
|
 |
S.Bastian,
M.J.Rekowski,
K.Witte,
D.M.Heckmann-Pohl,
and
F.Giffhorn
(2005).
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic efficiency.
|
| |
Appl Microbiol Biotechnol,
67,
654-663.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

