 |
PDBsum entry 2ign

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ign
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.10
- pyranose oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + O2 = 2-dehydro-D-glucose + H2O2
|
 |
 |
 |
 |
 |
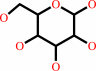 D-glucose
D-glucose
|
+
|
 O2
O2
|
=
|
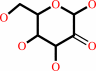 2-dehydro-D-glucose
2-dehydro-D-glucose
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:35104-35115
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for substrate binding and regioselective oxidation of monosaccharides at C3 by pyranose 2-oxidase.
|
|
M.Kujawa,
H.Ebner,
C.Leitner,
B.M.Hallberg,
M.Prongjit,
J.Sucharitakul,
R.Ludwig,
U.Rudsander,
C.Peterbauer,
P.Chaiyen,
D.Haltrich,
C.Divne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Pyranose 2-oxidase (P2Ox) participates in fungal lignin degradation by producing
the H2O2 needed for lignin-degrading peroxidases. The enzyme oxidizes cellulose-
and hemicellulose-derived aldopyranoses at C2 preferentially, but also on C3, to
the corresponding ketoaldoses. To investigate the structural determinants of
catalysis, covalent flavinylation, substrate binding, and regioselectivity,
wild-type and mutant P2Ox enzymes were produced and characterized biochemically
and structurally. Removal of the histidyl-FAD linkage resulted in a
catalytically competent enzyme containing tightly, but noncovalently bound FAD.
This mutant (H167A) is characterized by a 5-fold lower kcat, and a 35-mV lower
redox potential, although no significant structural changes were seen in its
crystal structure. In previous structures of P2Ox, the substrate loop (residues
452-457) covering the active site has been either disordered or in a
conformation incompatible with carbohydrate binding. We present here the crystal
structure of H167A in complex with a slow substrate, 2-fluoro-2-deoxy-D-glucose.
Based on the details of 2-fluoro-2-deoxy-D-glucose binding in position for
oxidation at C3, we also outline a probable binding mode for D-glucose
positioned for regioselective oxidation at C2. The tentative determinant for
discriminating between the two binding modes is the position of the O6 hydroxyl
group, which in the C2-oxidation mode can make favorable interactions with
Asp452 in the substrate loop and, possibly, a nearby arginine residue (Arg472).
We also substantiate our hypothesis with steady-state kinetics data for the
alanine replacements of Asp452 and Arg472 as well as the double alanine 452/472
mutant.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Details of ligand binding in nsP2Ox[ACT] and
His6-H167A[2FG]. Side-by-side view showing the active site in
the nsP2Ox[ACT] complex (A) and the His[6]-H167A[2FG] complex
(B). In B, the carbohydrate is positioned for oxidation at C3.
Firm interactions are noted for the C3 hydroxyl group and the
side chains of His^548 and Asn^593. The substrate loop is
highlighted in magenta. Atom-coloring scheme: carbon, beige
(protein), yellow (FAD), green (ligand); nitrogen, blue; oxygen,
red. C, superposition of the active sites in nsP2Ox[ACT] (beige)
and His[6]-H167A[2FG] (green).
|
 |
Figure 4.
FIGURE 4. Productive binding modes of D-glucose in the C2
and C3 orientations. A, observed binding of 2FG to the
His[6]-H167A[2FG] active site. Unbiased electron density for the
carbohydrate and part of the flavin ring. The electron density
was calculated using an early model where the ligand had not yet
been included. B, observed binding of 2FG in the C3-oxidation
orientation, and in the theoretical C2-oxidation orientation
(C). In C, the protein model from the His[6]-H167A[2FG] complex
was used for modeling the C2 orientation. The 2FG molecule was
rotated 180° about an axis defined roughly by a line running
through a point midway between the glucose atoms C5 and O5, and
a point midway between atoms C2 and C3. The same coloring scheme
is used as in Fig. 3.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
35104-35115)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Salaheddin,
Y.Takakura,
M.Tsunashima,
B.Stranzinger,
O.Spadiut,
M.Yamabhai,
C.K.Peterbauer,
and
D.Haltrich
(2010).
Characterisation of recombinant pyranose oxidase from the cultivated mycorrhizal basidiomycete Lyophyllum shimeji (hon-shimeji).
|
| |
Microb Cell Fact,
9,
57.
|
 |
|
|
|
|
 |
O.Spadiut,
G.Posch,
R.Ludwig,
D.Haltrich,
and
C.K.Peterbauer
(2010).
Evaluation of different expression systems for the heterologous expression of pyranose 2-oxidase from Trametes multicolor in E. coli.
|
| |
Microb Cell Fact,
9,
14.
|
 |
|
|
|
|
 |
O.Spadiut,
T.C.Tan,
I.Pisanelli,
D.Haltrich,
and
C.Divne
(2010).
Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
|
| |
FEBS J,
277,
2892-2909.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Orville,
G.T.Lountos,
S.Finnegan,
G.Gadda,
and
R.Prabhakar
(2009).
Crystallographic, spectroscopic, and computational analysis of a flavin C4a-oxygen adduct in choline oxidase.
|
| |
Biochemistry,
48,
720-728.
|
 |
|
|
|
|
 |
D.P.Heuts,
N.S.Scrutton,
W.S.McIntire,
and
M.W.Fraaije
(2009).
What's in a covalent bond? On the role and formation of covalently bound flavin cofactors.
|
| |
FEBS J,
276,
3405-3427.
|
 |
|
|
|
|
 |
I.Dreveny,
A.S.Andryushkova,
A.Glieder,
K.Gruber,
and
C.Kratky
(2009).
Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
|
| |
Biochemistry,
48,
3370-3377.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
C.Leitner,
C.Salaheddin,
B.Varga,
B.G.Vertessy,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design.
|
| |
FEBS J,
276,
776-792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Spadiut,
K.Radakovits,
I.Pisanelli,
C.Salaheddin,
M.Yamabhai,
T.C.Tan,
C.Divne,
and
D.Haltrich
(2009).
A thermostable triple mutant of pyranose 2-oxidase from Trametes multicolor with improved properties for biotechnological applications.
|
| |
Biotechnol J,
4,
525-534.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Wu,
P.M.Flatt,
H.Xu,
and
T.Mahmud
(2009).
Biosynthetic Gene Cluster of Cetoniacytone A, an Unusual Aminocyclitol from the Endosymbiotic Bacterium Actinomyces sp. Lu 9419.
|
| |
Chembiochem,
10,
304-314.
|
 |
|
|
|
|
 |
S.Nijvipakul,
J.Wongratana,
C.Suadee,
B.Entsch,
D.P.Ballou,
and
P.Chaiyen
(2008).
LuxG is a functioning flavin reductase for bacterial luminescence.
|
| |
J Bacteriol,
190,
1531-1538.
|
 |
|
|
|
|
 |
V.Joosten,
and
W.J.van Berkel
(2007).
Flavoenzymes.
|
| |
Curr Opin Chem Biol,
11,
195-202.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

