 |
PDBsum entry 1sb3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1sb3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 761 a.a.
761 a.a.
|
 |

|

|

|

|

|

|

|

 323 a.a.
323 a.a.
|
 |

|

|

|

|

|

|

|

 161 a.a.
161 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of 4-hydroxybenzoyl-coa reductase from thauera aromatica
|
|
Structure:
|
 |
4-hydroxybenzoyl-coa reductase alpha subunit. Chain: a, d. 4-hydroxybenzoyl-coa reductase beta subunit. Chain: b, e. 4-hydroxybenzoyl-coa reductase gamma subunit. Chain: c, f. Ec: 1.3.99.20
|
|
Source:
|
 |
Thauera aromatica. Organism_taxid: 59405. Strain: strain k, dsmz 6984. Strain: strain k, dsmz 6984
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.171
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
M.Unciuleac,E.Warkentin,C.C.Page,M.Boll,U.Ermler
|
Key ref:

|
 |
M.Unciuleac
et al.
(2004).
Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow.
Structure,
12,
2249-2256.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Feb-04
|
Release date:
|
21-Dec-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O33819
(HCRA_THAAR) -
4-hydroxybenzoyl-CoA reductase subunit alpha from Thauera aromatica
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
769 a.a.
761 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.1.7.1
- 4-hydroxybenzoyl-CoA reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxidized 2[4Fe-4S]-[ferredoxin] + benzoyl-CoA + H2O = 4-hydroxybenzoyl- CoA + reduced 2[4Fe-4S]-[ferredoxin] + 2 H+
|
 |
 |
 |
 |
 |
oxidized 2[4Fe-4S]-[ferredoxin]
Bound ligand (Het Group name = )
matches with 43.42% similarity
|
+
|
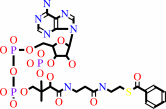 benzoyl-CoA
benzoyl-CoA
|
+
|
H2O
|
=
|
4-hydroxybenzoyl- CoA
|
+
|
reduced 2[4Fe-4S]-[ferredoxin]
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Flavin; Iron-sulfur; Mo cation
|
 |
 |
 |
 |
 |
 Flavin
Flavin
|
 Iron-sulfur
Iron-sulfur
|
Mo cation
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:2249-2256
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow.
|
|
M.Unciuleac,
E.Warkentin,
C.C.Page,
M.Boll,
U.Ermler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Mo-flavo-Fe/S-dependent heterohexameric protein complex 4-hydroxybenzoyl-CoA
reductase (4-HBCR, dehydroxylating) is a central enzyme of the anaerobic
degradation of phenolic compounds and belongs to the xanthine oxidase (XO)
family of molybdenum enzymes. Its X-ray structure was established at 1.6 A
resolution. The most pronounced difference between 4-HBCR and other structurally
characterized members of the XO family is the insertion of 40 amino acids within
cluster at a distance of
16.5 A to the isoalloxazine ring of FAD. The architecture of 4-HBCR and
concomitantly performed electron transfer rate calculations suggest an inverted
cluster to
the Mo over a distance of 55 A. The binding site of 4-hydroxybenzoyl-CoA is
located in an 18 A long channel lined up by several aromatic side chains around
the aromatic moiety, which are proposed to shield and stabilize the postulated
radical intermediates during catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Reaction Catalyzed by 4-Hydroxybenzoyl-CoA
Reductase

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
2249-2256)
copyright 2004.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Neumann,
and
S.Leimkühler
(2011).
The role of system-specific molecular chaperones in the maturation of molybdoenzymes in bacteria.
|
| |
Biochem Res Int,
2011,
850924.
|
 |
|
|
|
|
 |
M.Carmona,
M.T.Zamarro,
B.Blázquez,
G.Durante-Rodríguez,
J.F.Juárez,
J.A.Valderrama,
M.J.Barragán,
J.L.García,
and
E.Díaz
(2009).
Anaerobic catabolism of aromatic compounds: a genetic and genomic view.
|
| |
Microbiol Mol Biol Rev,
73,
71.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
G.Fuchs
(2008).
Anaerobic metabolism of aromatic compounds.
|
| |
Ann N Y Acad Sci,
1125,
82-99.
|
 |
|
|
|
|
 |
J.Johannes,
A.Bluschke,
N.Jehmlich,
M.von Bergen,
and
M.Boll
(2008).
Purification and characterization of active-site components of the putative p-cresol methylhydroxylase membrane complex from Geobacter metallireducens.
|
| |
J Bacteriol,
190,
6493-6500.
|
 |
|
|
|
|
 |
F.Peters,
D.Heintz,
J.Johannes,
A.van Dorsselaer,
and
M.Boll
(2007).
Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens.
|
| |
J Bacteriol,
189,
4729-4738.
|
 |
|
|
|
|
 |
C.D.Brondino,
M.J.Romão,
I.Moura,
and
J.J.Moura
(2006).
Molybdenum and tungsten enzymes: the xanthine oxidase family.
|
| |
Curr Opin Chem Biol,
10,
109-114.
|
 |
|
|
|
|
 |
W.Buckel,
and
B.T.Golding
(2006).
Radical enzymes in anaerobes.
|
| |
Annu Rev Microbiol,
60,
27-49.
|
 |
|
|
|
|
 |
M.Boll,
B.Schink,
A.Messerschmidt,
and
P.M.Kroneck
(2005).
Novel bacterial molybdenum and tungsten enzymes: three-dimensional structure, spectroscopy, and reaction mechanism.
|
| |
Biol Chem,
386,
999.
|
 |
|
|
|
|
 |
M.Boll,
and
G.Fuchs
(2005).
Unusual reactions involved in anaerobic metabolism of phenolic compounds.
|
| |
Biol Chem,
386,
989-997.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
| |

