 |
PDBsum entry 1qxo

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.3.5
- chorismate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-O-(1-carboxyvinyl)-3-phosphoshikimate = chorismate + phosphate
|
 |
 |
 |
 |
 |
5-O-(1-carboxyvinyl)-3-phosphoshikimate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
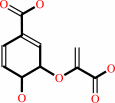 chorismate
chorismate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
11:1499-1511
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of chorismate synthase reveals a novel flavin binding site fundamental to a unique chemical reaction.
|
|
J.Maclean,
S.Ali.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of chorismate synthase (CS) from Streptococcus pneumoniae
has been solved to 2.0 A resolution in the presence of flavin mononucleotide
(FMN) and the substrate 5-enolpyruvyl-3-shikimate phosphate (EPSP). CS catalyses
the final step of the shikimate pathway and is a potential therapeutic target
for the rational design of novel antibacterials, antifungals, antiprotozoals,
and herbicides. CS is a tetramer with the monomer possessing a novel
beta-alpha-beta fold. The interactions between the enzyme, cofactor, and
substrate reveal the structural reasons underlying the unique catalytic
mechanism and identify the amino acids involved. This structure provides the
essential initial information necessary for the generation of novel
anti-infective compounds by a structure-guided medicinal chemistry approach.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. The Chorismate Synthase Active Site(A) Stereo
representation of interactions made by FMN in the closed form.
Active site residues are shown with green carbon atoms, FMN and
EPSP with gray carbon atoms. Red spheres represent conserved
water positions. Hydrogen bonds are shown as dashed lines. Some
residues shown are from an adjacent monomer and form
interactions that may stabilize the CS dimer. These residues are
shown in darker green, and are labeled with green text, while
residues from the monomer, which forms the majority of the
active site, have black labels.(B) Stereo representation of
interactions made by EPSP in the closed form. Active site
residues are shown with green carbon atoms, FMN and EPSP with
gray carbon atoms. Red spheres represent conserved water
positions. Hydrogen bonds are shown as dashed lines.(C) Stereo
diagram of the overlaid Ca traces of open (cyan ribbon) and
closed (green ribbon) forms, showing differences in the
conformations of loops L20 and L22. EPSP and FMN are shown with
gray carbon atoms. The side chains of residues His 110, Tyr 317,
Arg 337, Ser 338, and Asp 339 are shown for both forms (Open
form: cyan carbons, closed form: green carbons). L22 shows the
most significant movement, as demonstrated by the changes in the
position of Arg 337 and Ser 338. Tyr 317 on loop L20 adopts
different side chain conformations in open and closed forms, and
may have a role in maintaining the closed conformation of loop
L22. Hydrogen bond interactions between ligands and the
highlighted residues in the closed form are shown as dashed
lines. Corresponding interactions in the open form are omitted
for clarity.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2003,
11,
1499-1511)
copyright 2003.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.B.Barcellos,
R.A.Caceres,
and
W.F.de Azevedo
(2009).
Structural studies of shikimate dehydrogenase from Bacillus anthracis complexed with cofactor NADP.
|
| |
J Mol Model,
15,
147-155.
|
 |
|
|
|
|
 |
H.Unno,
S.Yamashita,
Y.Ikeda,
S.Y.Sekiguchi,
N.Yoshida,
T.Yoshimura,
M.Kusunoki,
T.Nakayama,
T.Nishino,
and
H.Hemmi
(2009).
New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
|
| |
J Biol Chem,
284,
9160-9167.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Ely,
J.E.Nunes,
E.K.Schroeder,
J.Frazzon,
M.S.Palma,
D.S.Santos,
and
L.A.Basso
(2008).
The Mycobacterium tuberculosis Rv2540c DNA sequence encodes a bifunctional chorismate synthase.
|
| |
BMC Biochem,
9,
13.
|
 |
|
|
|
|
 |
G.Rauch,
H.Ehammer,
S.Bornemann,
and
P.Macheroux
(2008).
Replacement of two invariant serine residues in chorismate synthase provides evidence that a proton relay system is essential for intermediate formation and catalytic activity.
|
| |
FEBS J,
275,
1464-1473.
|
 |
|
|
|
|
 |
H.Ehammer,
G.Rauch,
A.Prem,
B.Kappes,
and
P.Macheroux
(2007).
Conservation of NADPH utilization by chorismate synthase and its implications for the evolution of the shikimate pathway.
|
| |
Mol Microbiol,
65,
1249-1257.
|
 |
|
|
|
|
 |
S.O.Mansoorabadi,
C.J.Thibodeaux,
and
H.W.Liu
(2007).
The diverse roles of flavin coenzymes--nature's most versatile thespians.
|
| |
J Org Chem,
72,
6329-6342.
|
 |
|
|
|
|
 |
W.Kittleman,
C.J.Thibodeaux,
Y.N.Liu,
H.Zhang,
and
H.W.Liu
(2007).
Characterization and mechanistic studies of type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase from Staphylococcus aureus.
|
| |
Biochemistry,
46,
8401-8413.
|
 |
|
|
|
|
 |
P.Macheroux,
S.Ghisla,
C.Sanner,
H.Rüterjans,
and
F.Müller
(2005).
Reduced flavin: NMR investigation of N5-H exchange mechanism, estimation of ionisation constants and assessment of properties as biological catalyst.
|
| |
BMC Biochem,
6,
26.
|
 |
|
|
|
|
 |
K.Kitzing,
S.Auweter,
N.Amrhein,
and
P.Macheroux
(2004).
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site.
|
| |
J Biol Chem,
279,
9451-9461.
|
 |
|
|
|
|
 |
M.V.Dias,
F.Ely,
F.Canduri,
J.H.Pereira,
J.Frazzon,
L.A.Basso,
M.S.Palma,
W.F.de Azevedo,
and
D.S.Santos
(2004).
Crystallization and preliminary X-ray crystallographic analysis of chorismate synthase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2003-2005.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

