 |
PDBsum entry 1qr2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1qr2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.10.5.1
- ribosyldihydronicotinamide dehydrogenase (quinone).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide + a quinone + H+ = beta-nicotinamide D-riboside + a quinol
|
 |
 |
 |
 |
 |
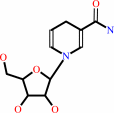 1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
1-(beta-D-ribofuranosyl)-1,4-dihydronicotinamide
|
+
|
 quinone
quinone
|
+
|
H(+)
|
=
|
beta-nicotinamide D-riboside
|
+
|
quinol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Zn(2+)
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
Zn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:9881-9886
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
|
|
C.E.Foster,
M.A.Bianchet,
P.Talalay,
Q.Zhao,
L.M.Amzel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In mammals, two separate but homologous cytosolic quinone reductases have been
identified: NAD(P)H:quinone oxidoreductase type 1 (QR1) (EC 1.6.99.2) and
quinone reductase type 2 (QR2). Although QR1 and QR2 are nearly 50% identical in
protein sequence, they display markedly different catalytic properties and
substrate specificities. We report here two crystal structures of QR2: in its
native form and bound to menadione (vitamin K(3)), a physiological substrate.
Phases were obtained by molecular replacement, using our previously determined
rat QR1 structure as the search model. QR2 shares the overall fold of the major
catalytic domain of QR1, but lacks the smaller C-terminal domain. The FAD
binding sites of QR1 and QR2 are very similar, but their hydride donor binding
sites are considerably different. Unexpectedly, we found that QR2 contains a
specific metal binding site, which is not present in QR1. Two histidine
nitrogens, one cysteine thiol, and a main chain carbonyl group are involved in
metal coordination. The metal binding site is solvent-accessible, and is
separated from the FAD cofactor by a distance of about 13 A.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Müller,
and
A.Hemphill
(2011).
Identification of a host cell target for the thiazolide class of broad-spectrum anti-parasitic drugs.
|
| |
Exp Parasitol,
128,
145-150.
|
 |
|
|
|
|
 |
V.Leclerc,
M.Ettaoussi,
M.Rami,
A.Farce,
J.A.Boutin,
P.Delagrange,
D.H.Caignard,
P.Renard,
P.Berthelot,
and
S.Yous
(2011).
Design and synthesis of naphthalenic derivatives as new ligands at the melatonin binding site MT3.
|
| |
Eur J Med Chem,
46,
1622-1629.
|
 |
|
|
|
|
 |
A.Maiti,
P.V.Reddy,
M.Sturdy,
L.Marler,
S.D.Pegan,
A.D.Mesecar,
J.M.Pezzuto,
and
M.Cushman
(2009).
Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
|
| |
J Med Chem,
52,
1873-1884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Winger,
O.Hantschel,
G.Superti-Furga,
and
J.Kuriyan
(2009).
The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2).
|
| |
BMC Struct Biol,
9,
7.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.W.Gaikwad,
L.Yang,
E.G.Rogan,
and
E.L.Cavalieri
(2009).
Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones.
|
| |
Free Radic Biol Med,
46,
253-262.
|
 |
|
|
|
|
 |
S.Sollner,
and
P.Macheroux
(2009).
New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation.
|
| |
FEBS J,
276,
4313-4324.
|
 |
|
|
|
|
 |
T.P.Roosild,
S.Castronovo,
S.Miller,
C.Li,
T.Rasmussen,
W.Bartlett,
B.Gunasekera,
S.Choe,
and
I.R.Booth
(2009).
KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation.
|
| |
Structure,
17,
893-903.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Ito,
M.Nakanishi,
W.C.Lee,
Y.Zhi,
H.Sasaki,
S.Zenno,
K.Saigo,
Y.Kitade,
and
M.Tanokura
(2008).
Expansion of substrate specificity and catalytic mechanism of azoreductase by X-ray crystallography and site-directed mutagenesis.
|
| |
J Biol Chem,
283,
13889-13896.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Fu,
L.Buryanovskyy,
and
Z.Zhang
(2008).
Quinone reductase 2 is a catechol quinone reductase.
|
| |
J Biol Chem,
283,
23829-23835.
|
 |
|
|
|
|
 |
M.M.AbuKhader,
J.Heap,
C.I.De Matteis,
S.W.Doughty,
N.Minton,
and
M.Paoli
(2007).
Crystallization and preliminary X-ray characterization of the Bacillus amyloliquefaciens YwrO enzyme.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
746-750.
|
 |
|
|
|
|
 |
S.Sollner,
R.Nebauer,
H.Ehammer,
A.Prem,
S.Deller,
B.A.Palfey,
G.Daum,
and
P.Macheroux
(2007).
Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification.
|
| |
FEBS J,
274,
1328-1339.
|
 |
|
|
|
|
 |
U.Rix,
O.Hantschel,
G.Dürnberger,
L.L.Remsing Rix,
M.Planyavsky,
N.V.Fernbach,
I.Kaupe,
K.L.Bennett,
P.Valent,
J.Colinge,
T.Köcher,
and
G.Superti-Furga
(2007).
Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets.
|
| |
Blood,
110,
4055-4063.
|
 |
|
|
|
|
 |
K.Ito,
M.Nakanishi,
W.C.Lee,
H.Sasaki,
S.Zenno,
K.Saigo,
Y.Kitade,
and
M.Tanokura
(2006).
Three-dimensional structure of AzoR from Escherichia coli. An oxidereductase conserved in microorganisms.
|
| |
J Biol Chem,
281,
20567-20576.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Wang,
and
A.K.Jaiswal
(2006).
Nuclear factor Nrf2 and antioxidant response element regulate NRH:quinone oxidoreductase 2 (NQO2) gene expression and antioxidant induction.
|
| |
Free Radic Biol Med,
40,
1119-1130.
|
 |
|
|
|
|
 |
A.E.Speers,
and
B.F.Cravatt
(2005).
A tandem orthogonal proteolysis strategy for high-content chemical proteomics.
|
| |
J Am Chem Soc,
127,
10018-10019.
|
 |
|
|
|
|
 |
J.A.Boutin,
F.Chatelain-Egger,
F.Vella,
P.Delagrange,
and
G.Ferry
(2005).
Quinone reductase 2 substrate specificity and inhibition pharmacology.
|
| |
Chem Biol Interact,
151,
213-228.
|
 |
|
|
|
|
 |
J.Gorman,
and
L.Shapiro
(2005).
Crystal structures of the tryptophan repressor binding protein WrbA and complexes with flavin mononucleotide.
|
| |
Protein Sci,
14,
3004-3012.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Iskander,
and
A.K.Jaiswal
(2005).
Quinone oxidoreductases in protection against myelogenous hyperplasia and benzene toxicity.
|
| |
Chem Biol Interact,
153,
147-157.
|
 |
|
|
|
|
 |
P.Talalay
(2005).
A fascination with enzymes: the journey not the arrival matters.
|
| |
J Biol Chem,
280,
28829-28847.
|
 |
|
|
|
|
 |
D.J.Long,
K.Iskander,
A.Gaikwad,
M.Arin,
D.R.Roop,
R.Knox,
R.Barrios,
and
A.K.Jaiswal
(2002).
Disruption of dihydronicotinamide riboside:quinone oxidoreductase 2 (NQO2) leads to myeloid hyperplasia of bone marrow and decreased sensitivity to menadione toxicity.
|
| |
J Biol Chem,
277,
46131-46139.
|
 |
|
|
|
|
 |
G.Cavelier,
and
L.M.Amzel
(2001).
Mechanism of NAD(P)H:quinone reductase: Ab initio studies of reduced flavin.
|
| |
Proteins,
43,
420-432.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
A.T.Dinkova-Kostova,
and
P.Talalay
(2000).
Persuasive evidence that quinone reductase type 1 (DT diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen.
|
| |
Free Radic Biol Med,
29,
231-240.
|
 |
|
|
|
|
 |
C.E.Foster,
M.A.Bianchet,
P.Talalay,
M.Faig,
and
L.M.Amzel
(2000).
Structures of mammalian cytosolic quinone reductases.
|
| |
Free Radic Biol Med,
29,
241-245.
|
 |
|
|
|
|
 |
S.Miller,
L.S.Ness,
C.M.Wood,
B.C.Fox,
and
I.R.Booth
(2000).
Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli.
|
| |
J Bacteriol,
182,
6536-6540.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

