 |
PDBsum entry 1v4b

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1v4b
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.6.5.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.7.1.17
- FMN-dependent NADH-azoreductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N,N-dimethyl-1,4-phenylenediamine + anthranilate + 2 NAD+ = 2-(4-dimethylaminophenyl)diazenylbenzoate + 2 NADH + 2 H+
|
 |
 |
 |
 |
 |
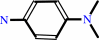 N,N-dimethyl-1,4-phenylenediamine
N,N-dimethyl-1,4-phenylenediamine
|
+
|
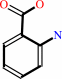 anthranilate
anthranilate
|
+
|
 2
×
NAD(+)
2
×
NAD(+)
|
=
|
2-(4-dimethylaminophenyl)diazenylbenzoate
|
+
|
 2
×
NADH
2
×
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:20567-20576
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of AzoR from Escherichia coli. An oxidereductase conserved in microorganisms.
|
|
K.Ito,
M.Nakanishi,
W.C.Lee,
H.Sasaki,
S.Zenno,
K.Saigo,
Y.Kitade,
M.Tanokura.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of AzoR (azoreductase) has been determined in complex with
FMN for two different crystal forms at 1.8 and 2.2 A resolution. AzoR is an
oxidoreductase isolated from Escherichia coli as a protein responsible for the
degradation of azo compounds. This enzyme is an FMN-dependent NADH-azoreductase
and catalyzes the reductive cleavage of azo groups by a ping-pong mechanism. The
structure suggests that AzoR acts in a homodimeric state forming the two
identical catalytic sites to which both monomers contribute. The structure
revealed that each monomer of AzoR has a flavodoxin-like structure, without the
explicit overall amino acid sequence homology. Superposition of the structures
from the two different crystal forms revealed the conformational change and
suggested a mechanism for accommodating substrates of different size.
Furthermore, comparison of the active site structure with that of NQO1 complexed
with substrates provides clues to the possible substrate-binding mechanism of
AzoR.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIGURE 2. Interactions between FMN and amino acid residues
in the active site. a, schematic diagram showing contacts of the
FMN cofactor to amino acid residues. Hydrogen bonds are shown as
broken green lines (red residue numbers) and van der Waals'
interactions by red shading (black residue numbers). Each atom
element is represented by a sphere of different colors with a
chemical symbol. b, SIGMAA-weighted 2mF[o] - DF[c] electron
density maps surrounding the FMN. The map was calculated using
the data of P4[2]2[1]2 crystal structure and is contoured at 1.2
 .
c, the electrostatic potential of AzoR is mapped onto the
solvent-accessible surface, as calculated with GRASP. The FMN
molecules shown in a and b are represented by a stick model,
with color coding identical to that in Fig. 1. .
c, the electrostatic potential of AzoR is mapped onto the
solvent-accessible surface, as calculated with GRASP. The FMN
molecules shown in a and b are represented by a stick model,
with color coding identical to that in Fig. 1.
|
 |
Figure 5.
FIGURE 5. Comparison of the active site of AzoR with that
of NQO1. One active site is shown for each of AzoR (a) and rat
NQO1 (b) (Protein Data Bank accession code 1QRD). The flavin
cofactors, duroquinone, and the side chains of conserved amino
acid residues in both enzymes are represented by a stick model,
with carbon atoms in gray, oxygen atoms in red, nitrogen atoms
in blue, and phosphorous atoms in orange. The polypeptide
moieties of one subunit for each enzyme are drawn as C  traces
in yellow and green. traces
in yellow and green.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
20567-20576)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Chen,
J.Feng,
O.Kweon,
H.Xu,
and
C.E.Cerniglia
(2010).
Identification and molecular characterization of a novel flavin-free NADPH preferred azoreductase encoded by azoB in Pigmentiphaga kullae K24.
|
| |
BMC Biochem,
11,
13.
|
 |
|
|
|
|
 |
S.Bürger,
and
A.Stolz
(2010).
Characterisation of the flavin-free oxygen-tolerant azoreductase from Xenophilus azovorans KF46F in comparison to flavin-containing azoreductases.
|
| |
Appl Microbiol Biotechnol,
87,
2067-2076.
|
 |
|
|
|
|
 |
G.Liu,
J.Zhou,
Q.S.Fu,
and
J.Wang
(2009).
The Escherichia coli azoreductase AzoR Is involved in resistance to thiol-specific stress caused by electrophilic quinones.
|
| |
J Bacteriol,
191,
6394-6400.
|
 |
|
|
|
|
 |
T.Ooi,
T.Shibata,
K.Matsumoto,
S.Kinoshita,
and
S.Taguchi
(2009).
Comparative enzymatic analysis of azoreductases from Bacillus sp. B29.
|
| |
Biosci Biotechnol Biochem,
73,
1209-1211.
|
 |
|
|
|
|
 |
G.Liu,
J.Zhou,
J.Wang,
B.Yan,
J.Li,
H.Lu,
Y.Qu,
and
R.Jin
(2008).
Site-directed mutagenesis of substrate binding sites of azoreductase from Rhodobacter sphaeroides.
|
| |
Biotechnol Lett,
30,
869-875.
|
 |
|
|
|
|
 |
H.Chen,
H.Xu,
O.Kweon,
S.Chen,
and
C.E.Cerniglia
(2008).
Functional role of Trp-105 of Enterococcus faecalis azoreductase (AzoA) as resolved by structural and mutational analysis.
|
| |
Microbiology,
154,
2659-2667.
|
 |
|
|
|
|
 |
V.V.Dawkar,
U.U.Jadhav,
S.U.Jadhav,
and
S.P.Govindwar
(2008).
Biodegradation of disperse textile dye Brown 3REL by newly isolated Bacillus sp. VUS.
|
| |
J Appl Microbiol,
105,
14-24.
|
 |
|
|
|
|
 |
M.Roldo,
E.Barbu,
J.F.Brown,
D.W.Laight,
J.D.Smart,
and
J.Tsibouklis
(2007).
Azo compounds in colon-specific drug delivery.
|
| |
Expert Opin Drug Deliv,
4,
547-560.
|
 |
|
|
|
|
 |
T.Ooi,
T.Shibata,
R.Sato,
H.Ohno,
S.Kinoshita,
T.L.Thuoc,
and
S.Taguchi
(2007).
An azoreductase, aerobic NADH-dependent flavoprotein discovered from Bacillus sp.: functional expression and enzymatic characterization.
|
| |
Appl Microbiol Biotechnol,
75,
377-386.
|
 |
|
|
|
|
 |
Y.Nishiya,
and
Y.Yamamoto
(2007).
Characterization of a NADH:dichloroindophenol oxidoreductase from Bacillus subtilis.
|
| |
Biosci Biotechnol Biochem,
71,
611-614.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

