
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.4.1.1.22
- histidine decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-histidine + H+ = histamine + CO2
|
 |
 |
 |
 |
 |
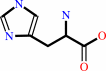 L-histidine
L-histidine
|
+
|
H(+)
|
=
|
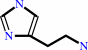 histamine
histamine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyruvate or pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyruvate
Pyruvate
|
or
|
 pyridoxal 5'-phosphate
pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
230:516-528
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Refined structure of the pyruvoyl-dependent histidine decarboxylase from Lactobacillus 30a.
|
|
T.Gallagher,
D.A.Rozwarski,
S.R.Ernst,
M.L.Hackert.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the pyruvoyl-dependent histidine decarboxylase from
Lactobacillus 30a has been refined to an R-value of 0.15 (for the 5.0 to 2.5 A
resolution shell) and 0.17 (for the 10.0 to 2.5 A resolution shell). A
description of the overall structure is presented, focusing on secondary
structure and subunit association. The enzyme is a hexamer of alpha beta
subunits. Separate alpha and beta-chains arise from an autocatalytic cleavage
reaction between two serine residues, which results in the pyruvoyl cofactor.
The central core of the alpha beta subunit is a beta-sandwich which consists of
two face-to-face three-stranded antiparallel beta-sheets, flanked by
alpha-helices on each side. The beta-sandwich creates a stable fold that allows
conformational strain to be introduced across an internal cleavage region
between the alpha and beta chains and places the pyruvoyl cofactor in a position
for efficient electron withdrawal from the substrate. Three alpha beta subunits
are related by a molecular three-fold symmetry axis to form a trimer whose
interfaces have complementary surfaces and extensive molecular interactions.
Each of the interfaces contains an active site and a solvent channel that leads
from the active site to the exterior of the molecule. The trimers are related by
a crystallographic two-fold symmetry axis to form the hexamer with an overall
dumbbell shape. The interface between trimers has few molecular interactions.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.J.Yokoi,
Y.Harada,
K.Shozen,
M.Satomi,
A.Taketo,
and
K.Kodaira
(2011).
Characterization of the histidine decarboxylase gene of Staphylococcus epidermidis TYH1 coded on the staphylococcal cassette chromosome.
|
| |
Gene,
477,
32-41.
|
 |
|
|
|
|
 |
S.C.Davis,
S.Clark,
J.R.Hayes,
T.L.Green,
and
C.A.Gruetter
(2011).
Up-regulation of histidine decarboxylase expression and histamine content in B16F10 murine melanoma cells.
|
| |
Inflamm Res,
60,
55-61.
|
 |
|
|
|
|
 |
S.Bale,
K.Baba,
D.E.McCloskey,
A.E.Pegg,
and
S.E.Ealick
(2010).
Complexes of Thermotoga maritimaS-adenosylmethionine decarboxylase provide insights into substrate specificity.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
181-189.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Bale,
and
S.E.Ealick
(2010).
Structural biology of S-adenosylmethionine decarboxylase.
|
| |
Amino Acids,
38,
451-460.
|
 |
|
|
|
|
 |
J.Crugeiras,
A.Rios,
E.Riveiros,
T.L.Amyes,
and
J.P.Richard
(2008).
Glycine enolates: the effect of formation of iminium ions to simple ketones on alpha-amino carbon acidity and a comparison with pyridoxal iminium ions.
|
| |
J Am Chem Soc,
130,
2041-2050.
|
 |
|
|
|
|
 |
P.M.Lucas,
W.A.Wolken,
O.Claisse,
J.S.Lolkema,
and
A.Lonvaud-Funel
(2005).
Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006.
|
| |
Appl Environ Microbiol,
71,
1417-1424.
|
 |
|
|
|
|
 |
A.Barzilai,
S.Kumar,
H.Wolfson,
and
R.Nussinov
(2004).
Potential folding-function interrelationship in proteins.
|
| |
Proteins,
56,
635-649.
|
 |
|
|
|
|
 |
A.V.Toms,
C.Kinsland,
D.E.McCloskey,
A.E.Pegg,
and
S.E.Ealick
(2004).
Evolutionary links as revealed by the structure of Thermotoga maritima S-adenosylmethionine decarboxylase.
|
| |
J Biol Chem,
279,
33837-33846.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Schmitzberger,
M.L.Kilkenny,
C.M.Lobley,
M.E.Webb,
M.Vinkovic,
D.Matak-Vinkovic,
M.Witty,
D.Y.Chirgadze,
A.G.Smith,
C.Abell,
and
T.L.Blundell
(2003).
Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase.
|
| |
EMBO J,
22,
6193-6204.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Worley,
E.Schelp,
A.F.Monzingo,
S.Ernst,
and
J.D.Robertus
(2002).
Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
|
| |
Proteins,
46,
321-329.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Poppe
(2001).
Methylidene-imidazolone: a novel electrophile for substrate activation.
|
| |
Curr Opin Chem Biol,
5,
512-524.
|
 |
|
|
|
|
 |
F.S.Tahanejad,
H.Naderi-Manesh,
B.Habibinejad,
and
M.Mahmoudian
(2000).
Homology-based molecular modelling of PLP-dependent histidine decarboxylase from Mmorganella morganii.
|
| |
Eur J Med Chem,
35,
567-576.
|
 |
|
|
|
|
 |
H.Paulus
(2000).
Protein splicing and related forms of protein autoprocessing.
|
| |
Annu Rev Biochem,
69,
447-496.
|
 |
|
|
|
|
 |
N.M.Okeley,
and
W.A.van der Donk
(2000).
Novel cofactors via post-translational modifications of enzyme active sites.
|
| |
Chem Biol,
7,
R159-R171.
|
 |
|
|
|
|
 |
H.Xiong,
and
A.E.Pegg
(1999).
Mechanistic studies of the processing of human S-adenosylmethionine decarboxylase proenzyme. Isolation of an ester intermediate.
|
| |
J Biol Chem,
274,
35059-35066.
|
 |
|
|
|
|
 |
J.L.Ekstrom,
I.I.Mathews,
B.A.Stanley,
A.E.Pegg,
and
S.E.Ealick
(1999).
The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold.
|
| |
Structure,
7,
583-595.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Xu,
D.Buckley,
C.Guan,
and
H.C.Guo
(1999).
Structural insights into the mechanism of intramolecular proteolysis.
|
| |
Cell,
98,
651-661.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.B.Perler
(1998).
Breaking up is easy with esters.
|
| |
Nat Struct Biol,
5,
249-252.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
H.Xiong,
B.A.Stanley,
B.L.Tekwani,
and
A.E.Pegg
(1997).
Processing of mammalian and plant S-adenosylmethionine decarboxylase proenzymes.
|
| |
J Biol Chem,
272,
28342-28348.
|
 |
|
|
|
|
 |
T.W.Hamelryck,
M.H.Dao-Thi,
F.Poortmans,
M.J.Chrispeels,
L.Wyns,
and
R.Loris
(1996).
The crystallographic structure of phytohemagglutinin-L.
|
| |
J Biol Chem,
271,
20479-20485.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

