 |
PDBsum entry 1pg8

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of l-methionine alpha-, gamma-lyase
|
|
Structure:
|
 |
Methionine gamma-lyase. Chain: a, b, c, d. Synonym: l-methioninase. Ec: 4.4.1.11
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303
|
|
Biol. unit:
|
 |
 Octamer (from
)
Octamer (from
)
|
|
Resolution:
|
 |
|
2.68Å
|
R-factor:
|
0.182
|
R-free:
|
0.236
|
|
|
Authors:
|
 |
T.W.Allen,V.Sridhar,G.S.Prasad,Q.Han,M.Xu,Y.Tan,R.M.Hoffman, S.Ramaswamy
|
|
Date:
|
 |
|
28-May-03
|
Release date:
|
08-Jun-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P13254
(MEGL_PSEPU) -
L-methionine gamma-lyase from Pseudomonas putida
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
398 a.a.
398 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.4.1.11
- methionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionine + H2O = methanethiol + 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
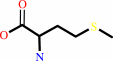 L-methionine
L-methionine
|
+
|
H2O
|
=
|
methanethiol
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
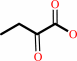 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.2
- homocysteine desulfhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + H2O = 2-oxobutanoate + hydrogen sulfide + NH4+ + H+
|
 |
 |
 |
 |
 |
 L-homocysteine
L-homocysteine
|
+
|
H2O
|
=
|
2-oxobutanoate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
hydrogen sulfide
|
+
|
NH4(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

