 |
PDBsum entry 1ok4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate
|
|
Structure:
|
 |
Fructose-bisphosphate aldolase class i. Chain: a, b, c, d, e, f, g, h, i, j. Synonym: fbp aldolase. Engineered: yes. Other_details: lys177 schiff-base with dhap
|
|
Source:
|
 |
Thermoproteus tenax. Organism_taxid: 2271. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Pentamer (from PDB file)
Pentamer (from PDB file)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.164
|
R-free:
|
0.185
|
|
|
Authors:
|
 |
E.Lorentzen,P.Zwart,A.Stark,R.Hensel,B.Siebers,E.Pohl
|
Key ref:

|
 |
E.Lorentzen
et al.
(2003).
Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J Biol Chem,
278,
47253-47260.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Jul-03
|
Release date:
|
04-Sep-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P58315
(ALF1_THETK) -
Fructose-bisphosphate aldolase class 1 from Thermoproteus tenax (strain ATCC 35583 / DSM 2078 / JCM 9277 / NBRC 100435 / Kra 1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
263 a.a.
251 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
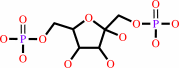 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 90.00% similarity
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:47253-47260
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
|
|
E.Lorentzen,
E.Pohl,
P.Zwart,
A.Stark,
R.B.Russell,
T.Knura,
R.Hensel,
B.Siebers.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fructose-1,6-bisphosphate aldolase (FBPA) catalyzes the reversible cleavage of
fructose 1,6-bisphosphate to glyceraldehyde 3-phosphate and dihydroxyacetone
phosphate in the glycolytic pathway. FBPAs from archaeal organisms have recently
been identified and characterized as a divergent family of proteins. Here, we
report the first crystal structure of an archaeal FBPA at 1.9-A resolution. The
structure of this 280-kDa protein complex was determined using single wavelength
anomalous dispersion followed by 10-fold non-crystallographic symmetry averaging
and refined to an R-factor of 14.9% (Rfree 17.9%). The protein forms a dimer of
pentamers, consisting of subunits adopting the ubiquitous (betaalpha)8 barrel
fold. Additionally, a crystal structure of the archaeal FBPA covalently bound to
dihydroxyacetone phosphate was solved at 2.1-A resolution. Comparison of the
active site residues with those of classical FBPAs, which share no significant
sequence identity but display the same overall fold, reveals a common ancestry
between these two families of FBPAs. Structural comparisons, furthermore,
establish an evolutionary link to the triosephosphate isomerases, a superfamily
hitherto considered independent from the superfamily of aldolases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. A, ribbon diagram of the crystal structure of the
Tt-FBPA decamer. Two pentamers face each other with the
N-terminal side of the TIM barrels to form the decamer. Each
monomer in the pentamers is shown in different colors and the
two pentamers in different shadings. The active sites are
located at the C termini of the TIM barrels and point away from
the pentamer-pentamer interface. B, one pentamer is shown in a
view perpendicular to that of A, using the same color coding.
|
 |
Figure 3.
FIG. 3. Close-up of the central hole of the pentamer in the
same orientation and color coding as Fig. 1B. The residues
Phe-119, Trp-121, and Lys-122 from each monomer, shown in
ball-and-stick representation, point into the central hole of
the pentamer and perform a continuous stacking.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
47253-47260)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Du,
R.F.Say,
W.Lü,
G.Fuchs,
and
O.Einsle
(2011).
Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
|
| |
Nature,
478,
534-537.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Diaz,
K.B.Xavier,
and
S.T.Miller
(2009).
The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
|
| |
PLoS One,
4,
e6820.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Samland,
M.Wang,
and
G.A.Sprenger
(2008).
MJ0400 from Methanocaldococcus jannaschii exhibits fructose-1,6-bisphosphate aldolase activity.
|
| |
FEMS Microbiol Lett,
281,
36-41.
|
 |
|
|
|
|
 |
A.Pauluhn,
H.Ahmed,
E.Lorentzen,
S.Buchinger,
D.Schomburg,
B.Siebers,
and
E.Pohl
(2008).
Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax.
|
| |
Proteins,
72,
35-43.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Zaparty,
B.Tjaden,
R.Hensel,
and
B.Siebers
(2008).
The central carbohydrate metabolism of the hyperthermophilic crenarchaeote Thermoproteus tenax: pathways and insights into their regulation.
|
| |
Arch Microbiol,
190,
231-245.
|
 |
|
|
|
|
 |
N.N.Smith,
and
D.T.Gallagher
(2008).
Structure and lability of archaeal dehydroquinase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
886-892.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.K.Samland,
and
G.A.Sprenger
(2006).
Microbial aldolases as C-C bonding enzymes--unknown treasures and new developments.
|
| |
Appl Microbiol Biotechnol,
71,
253-264.
|
 |
|
|
|
|
 |
V.F.Waingeh,
C.D.Gustafson,
E.I.Kozliak,
S.L.Lowe,
H.R.Knull,
and
K.A.Thomasson
(2006).
Glycolytic enzyme interactions with yeast and skeletal muscle F-actin.
|
| |
Biophys J,
90,
1371-1384.
|
 |
|
|
|
|
 |
B.Siebers,
and
P.Schönheit
(2005).
Unusual pathways and enzymes of central carbohydrate metabolism in Archaea.
|
| |
Curr Opin Microbiol,
8,
695-705.
|
 |
|
|
|
|
 |
J.Aishima,
D.S.Russel,
L.J.Guibas,
P.D.Adams,
and
A.T.Brunger
(2005).
Automated crystallographic ligand building using the medial axis transform of an electron-density isosurface.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1354-1363.
|
 |
|
|
|
|
 |
A.Stark,
A.Shkumatov,
and
R.B.Russell
(2004).
Finding functional sites in structural genomics proteins.
|
| |
Structure,
12,
1405-1412.
|
 |
|
|
|
|
 |
B.Liotard,
and
J.Sygusch
(2004).
Purification, crystallization and preliminary X-ray analysis of native and selenomethionine class I tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
528-530.
|
 |
|
|
|
|
 |
B.Siebers,
B.Tjaden,
K.Michalke,
C.Dörr,
H.Ahmed,
M.Zaparty,
P.Gordon,
C.W.Sensen,
A.Zibat,
H.P.Klenk,
S.C.Schuster,
and
R.Hensel
(2004).
Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data.
|
| |
J Bacteriol,
186,
2179-2194.
|
 |
|
|
|
|
 |
P.H.Zwart,
G.G.Langer,
and
V.S.Lamzin
(2004).
Modelling bound ligands in protein crystal structures.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2230-2239.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

