 |
PDBsum entry 1oat

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Aminotransferase
|
PDB id
|
|

|

|
1oat
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.13
- ornithine aminotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2-oxocarboxylate + L-ornithine = L-glutamate 5-semialdehyde + an L-alpha-amino acid
|
 |
 |
 |
 |
 |
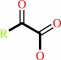 2-oxocarboxylate
2-oxocarboxylate
|
+
|
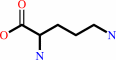 L-ornithine
L-ornithine
|
=
|
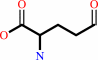 L-glutamate 5-semialdehyde
L-glutamate 5-semialdehyde
|
+
|
L-alpha-amino acid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
277:81
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human recombinant ornithine aminotransferase.
|
|
B.W.Shen,
M.Hennig,
E.Hohenester,
J.N.Jansonius,
T.Schirmer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ornithine aminotransferase (OAT), a pyridoxal-5'-phosphate dependent enzyme,
catalyses the transfer of the delta-amino group of L-ornithine to
2-oxoglutarate, producing L-glutamate-gamma-semialdehyde, which spontaneously
cyclizes to pyrroline-5-carboxylate, and L-glutamate. The crystal structure
determination of human recombinant OAT is described in this paper. As a first
step, the structure was determined at low resolution (6 A) by molecular
replacement using the refined structure of dialkylglycine decarboxylase as a
search model. Crystallographic phases were then refined and extended in a
step-wise fashion to 2.5 A by cyclic averaging of the electron density
corresponding to the three monomers within the asymmetric unit. Interpretation
of the resulting map was straightforward and refinement of the model resulted in
an R-factor of 17.1% (Rfree=24.3%). The success of the procedure demonstrates
the power of real-space molecular averaging even with only threefold redundancy.
The alpha6-hexameric molecule is a trimer of intimate dimers with a
monomer-monomer interface of 5500 A2 per subunit. The three dimers are related
by an approximate 3-fold screw axis with a translational component of 18 A. The
monomer fold is that of a typical representative of subgroup 2 aminotransferases
and very similar to those described for dialkylglycine decarboxylase from
Pseudomonas cepacia and glutamate-1-semialdehyde aminomutase from Synechococcus.
It consists of a large domain that contributes most to the subunit interface, a
C-terminal small domain most distant to the 2-fold axis and an N-terminal region
that contains a helix, a loop and a three stranded beta-meander embracing a
protrusion in the large domain of the second subunit of the dimer. The large
domain contains the characteristic central seven-stranded beta-sheet (agfedbc)
covered by eight helices in a typical alpha/beta fold. The cofactor
pyridoxal-5'-phosphate is bound through a Schiff base to Lys292, located in the
loop between strands f and g. The C-terminal domain includes a four-stranded
antiparallel beta-sheet in contact with the large domain and three further
helices at the far end of the subunit. The active sites of the dimer lie, about
25 A apart, at the subunit and domain interfaces. The conical entrances are on
opposite sides of the dimer. In the active site, R180, E235 and R413 are
probable substrate binding residues. Structure-based sequence comparisons with
related transaminases in this work support that view. In patients suffering from
gyrate atrophy, a recessive hereditary genetic disorder that can cause blindness
in humans, ornithine aminotransferase activity is lacking. A large number of
frameshift and point mutations in the ornithine aminotransferase gene have been
identified in such patients. Possible effects of the various point mutations on
the structural stability or the catalytic competence of the enzyme are discussed
in light of the three-dimensional structure.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 8.
Figure 8. Molecular surface representation of the hexameric
structure of OAT. The monomers (shown in different colors and
labeled) form three tight dimers (AB, CC′, A′B′) that are
related by a pseudo-3-fold screw axis with a translation
component of 18 Å. The screw-axis and the three molecular
dyads are shown in magenta. (a) View along the screw axis. The
vertical dyad is crystallographic and relates C with C′ as
well as dimer AB with dimer A′B′. The two other dyads are
non-crystallographic. (b) View along the crystallographic dyad
and perpendicular to the pseudo-3-fold screw axis. The figures
were produced using program GRASP [Nicholls et al 1991].
|
 |
Figure 10.
Figure 10. Stereo view of the superimposed C^α-traces of
the refined monomer models of OAT (black) and DGD (red). The
molecular dyad is shown in green. The large insertion in the DGD
structure (loops at the lower-left) is responsible for tetramer
formation [Toney et al 1995a]. The Figure was produced using
GRASP [Nicholls et al 1991].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1998,
277,
81-0)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.H.Park,
R.Mirabella,
P.A.Bronstein,
G.M.Preston,
M.A.Haring,
C.K.Lim,
A.Collmer,
and
R.C.Schuurink
(2010).
Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence.
|
| |
Plant J,
64,
318-330.
|
 |
|
|
|
|
 |
J.J.Tanner
(2008).
Structural biology of proline catabolism.
|
| |
Amino Acids,
35,
719-730.
|
 |
|
|
|
|
 |
J.Stránská,
D.Kopecný,
M.Tylichová,
J.Snégaroff,
and
M.Sebela
(2008).
Ornithine delta-aminotransferase: An enzyme implicated in salt tolerance in higher plants.
|
| |
Plant Signal Behav,
3,
929-935.
|
 |
|
|
|
|
 |
V.Rajaram,
P.Ratna Prasuna,
H.S.Savithri,
and
M.R.Murthy
(2008).
Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding.
|
| |
Proteins,
70,
429-441.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Tahlan,
C.Anders,
A.Wong,
R.H.Mosher,
P.H.Beatty,
M.J.Brumlik,
A.Griffin,
C.Hughes,
J.Griffin,
B.Barton,
and
S.E.Jensen
(2007).
5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus.
|
| |
Chem Biol,
14,
131-142.
|
 |
|
|
|
|
 |
S.M.Tripathi,
and
R.Ramachandran
(2006).
Overexpression, purification and crystallization of lysine epsilon-aminotransferase (Rv3290c) from Mycobacterium tuberculosis H37Rv.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
572-575.
|
 |
|
|
|
|
 |
V.Rajaram,
K.Prasad,
P.Ratna Prasuna,
N.Ramachandra,
S.R.Bharath,
H.S.Savithri,
and
M.R.Murthy
(2006).
Cloning, purification, crystallization and preliminary X-ray crystallographic analysis of the biosynthetic N-acetylornithine aminotransferases from Salmonella typhimurium and Escherichia coli.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
980-983.
|
 |
|
|
|
|
 |
L.Naranjo,
M.Lamas-Maceiras,
R.V.Ullán,
S.Campoy,
F.Teijeira,
J.Casqueiro,
and
J.F.Martín
(2005).
Characterization of the oat1 gene of Penicillium chrysogenum encoding an omega-aminotransferase: induction by L-lysine, L-ornithine and L-arginine and repression by ammonium.
|
| |
Mol Genet Genomics,
274,
283-294.
|
 |
|
|
|
|
 |
M.Markova,
C.Peneff,
M.J.Hewlins,
T.Schirmer,
and
R.A.John
(2005).
Determinants of substrate specificity in omega-aminotransferases.
|
| |
J Biol Chem,
280,
36409-36416.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Storici,
D.De Biase,
F.Bossa,
S.Bruno,
A.Mozzarelli,
C.Peneff,
R.B.Silverman,
and
T.Schirmer
(2004).
Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5'-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and with the antiepilepsy drug vigabatrin.
|
| |
J Biol Chem,
279,
363-373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Kongsaeree,
C.Samanchart,
P.Laowanapiban,
S.Wiyakrutta,
and
V.Meevootisom
(2003).
Crystallization and preliminary X-ray crystallographic analysis of d-phenylglycine aminotransferase from Pseudomonas stutzeri ST201.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
953-954.
|
 |
|
|
|
|
 |
H.Kagamiyama,
and
H.Hayashi
(2001).
Release of enzyme strain during catalysis reduces the activation energy barrier.
|
| |
Chem Rec,
1,
385-394.
|
 |
|
|
|
|
 |
K.Soda,
T.Yoshimura,
and
N.Esaki
(2001).
Stereospecificity for the hydrogen transfer of pyridoxal enzyme reactions.
|
| |
Chem Rec,
1,
373-384.
|
 |
|
|
|
|
 |
G.Schneider,
H.Käck,
and
Y.Lindqvist
(2000).
The manifold of vitamin B6 dependent enzymes.
|
| |
Structure,
8,
R1-R6.
|
 |
|
|
|
|
 |
A.D.Kern,
M.A.Oliveira,
P.Coffino,
and
M.L.Hackert
(1999).
Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
|
| |
Structure,
7,
567-581.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.N.Scarsdale,
G.Kazanina,
S.Radaev,
V.Schirch,
and
H.T.Wright
(1999).
Crystal structure of rabbit cytosolic serine hydroxymethyltransferase at 2.8 A resolution: mechanistic implications.
|
| |
Biochemistry,
38,
8347-8358.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Storici,
G.Capitani,
D.De Biase,
M.Moser,
R.A.John,
J.N.Jansonius,
and
T.Schirmer
(1999).
Crystal structure of GABA-aminotransferase, a target for antiepileptic drug therapy.
|
| |
Biochemistry,
38,
8628-8634.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.G.Mashima,
R.G.Weleber,
N.G.Kennaway,
and
G.Inana
(1999).
Genotype-phenotype correlation of a pyridoxine-responsive form of gyrate atrophy.
|
| |
Ophthalmic Genet,
20,
219-224.
|
 |
|
|
|
|
 |
H.Hayashi,
H.Mizuguchi,
and
H.Kagamiyama
(1998).
The imine-pyridine torsion of the pyridoxal 5'-phosphate Schiff base of aspartate aminotransferase lowers its pKa in the unliganded enzyme and is crucial for the successive increase in the pKa during catalysis.
|
| |
Biochemistry,
37,
15076-15085.
|
 |
|
|
|
|
 |
J.N.Jansonius
(1998).
Structure, evolution and action of vitamin B6-dependent enzymes.
|
| |
Curr Opin Struct Biol,
8,
759-769.
|
 |
|
|
|
|
 |
S.Ishii,
H.Hayashi,
A.Okamoto,
and
H.Kagamiyama
(1998).
Aromatic L-amino acid decarboxylase: conformational change in the flexible region around Arg334 is required during the transaldimination process.
|
| |
Protein Sci,
7,
1802-1810.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

