 |
PDBsum entry 1ohy

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
4-aminobutyrate-aminotransferase inactivated by gamma-ethynyl gaba
|
|
Structure:
|
 |
4-aminobutyrate aminotransferase. Chain: a, b, c, d. Synonym: mitochondrial precursor, gamma-amino-n-butyrate transaminase, gaba transaminase, gaba aminotransferase, gaba-at, gaba-t, abat, gabat. Other_details: after reaction with 4-amino-5-hexynoic acid (geg) (gamma-ethynyl gaba) the fragment geg has been generated which is covalently attached via a double bond to c4a of plp and via a single bond to nz of the active site lys 329
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823. Organ: liver
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.198
|
R-free:
|
0.229
|
|
|
Authors:
|
 |
P.Storici,T.Schirmer
|
Key ref:

|
 |
P.Storici
et al.
(2004).
Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5'-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and with the antiepilepsy drug vigabatrin.
J Biol Chem,
279,
363-373.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
03-Jun-03
|
Release date:
|
16-Oct-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P80147
(GABT_PIG) -
4-aminobutyrate aminotransferase, mitochondrial from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
500 a.a.
461 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.19
- 4-aminobutyrate--2-oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-aminobutanoate + 2-oxoglutarate = succinate semialdehyde + L-glutamate
|
 |
 |
 |
 |
 |
4-aminobutanoate
Bound ligand (Het Group name = )
matches with 77.78% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
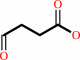 succinate semialdehyde
succinate semialdehyde
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.22
- (S)-3-amino-2-methylpropionate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-3-amino-2-methylpropanoate + 2-oxoglutarate = 2-methyl-3- oxopropanoate + L-glutamate
|
 |
 |
 |
 |
 |
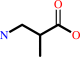 (S)-3-amino-2-methylpropanoate
(S)-3-amino-2-methylpropanoate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
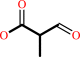 2-methyl-3- oxopropanoate
2-methyl-3- oxopropanoate
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 72.73% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:363-373
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5'-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and with the antiepilepsy drug vigabatrin.
|
|
P.Storici,
D.De Biase,
F.Bossa,
S.Bruno,
A.Mozzarelli,
C.Peneff,
R.B.Silverman,
T.Schirmer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Gamma-aminobutyric acid aminotransferase (GABA-AT) is a pyridoxal
5'-phosphate-dependent enzyme responsible for the degradation of the inhibitory
neurotransmitter GABA. GABA-AT is a validated target for antiepilepsy drugs
because its selective inhibition raises GABA concentrations in brain. The
antiepilepsy drug, gamma-vinyl-GABA (vigabatrin) has been investigated in the
past by various biochemical methods and resulted in several proposals for its
mechanisms of inactivation. In this study we solved and compared the crystal
structures of pig liver GABA-AT in its native form (to 2.3-A resolution) and in
complex with vigabatrin as well as with the close analogue gamma-ethynyl-GABA
(to 2.3 and 2.8 A, respectively). Both inactivators form a covalent ternary
adduct with the active site Lys-329 and the pyridoxal 5'-phosphate (PLP)
cofactor. The crystal structures provide direct support for specific
inactivation mechanisms proposed earlier on the basis of radio-labeling
experiments. The reactivity of GABA-AT crystals with the two GABA analogues was
also investigated by polarized absorption microspectrophotometry. The spectral
data are discussed in relation to the proposed mechanism. Intriguingly, all
cluster of yet unknown function at the
center of the dimeric molecule in the vicinity of the PLP cofactors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Stereographic projection of the active site of
GABA-AT. The final model is shown together with the 2F[o] - F[c]
map (contour level, 1.2  ). An acetate molecule
is found close to Arg-192, i.e. at a position where the
carboxylate moiety of the natural substrate GABA is expected to
bind (6). ). An acetate molecule
is found close to Arg-192, i.e. at a position where the
carboxylate moiety of the natural substrate GABA is expected to
bind (6).
|
 |
Figure 3.
FIG. 3. The [2Fe-2S] cluster at the center of the GABA-AT
dimer. A, structure of the GABA-AT dimer. The view is
approximately along the molecular 2-fold symmetry axis. The C
 traces of the two
subunits are shown in black and red. Helix 5 and its symmetry
mate are highlighted by thick gray traces. The [2Fe-2S] cluster
on the molecular 2-fold symmetry axis together with the
liganding cysteines and the two symmetry related PLP cofactors
are shown in full view. B, close-up view of A. The C traces of the two
subunits are shown in black and red. Helix 5 and its symmetry
mate are highlighted by thick gray traces. The [2Fe-2S] cluster
on the molecular 2-fold symmetry axis together with the
liganding cysteines and the two symmetry related PLP cofactors
are shown in full view. B, close-up view of A. The C  traces
have been omitted for clarity. Symmetry-related residues are
marked with the symbol "#." C, stereographic close-up view. The
molecular 2-fold axis is approximately along the vertical
direction. The native 2F[o] - F[c] omit map (magenta; contour
level 1.2 traces
have been omitted for clarity. Symmetry-related residues are
marked with the symbol "#." C, stereographic close-up view. The
molecular 2-fold axis is approximately along the vertical
direction. The native 2F[o] - F[c] omit map (magenta; contour
level 1.2  ) and the anomalous
difference map (light blue; contour level 4.5 ) and the anomalous
difference map (light blue; contour level 4.5  ) were
computed with data to 2.3-Å resolution (data set of the
vigabatrin complex). The iron and sulfur atoms were not included
for phasing. ) were
computed with data to 2.3-Å resolution (data set of the
vigabatrin complex). The iron and sulfur atoms were not included
for phasing.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
363-373)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.J.Headd,
N.Echols,
P.V.Afonine,
R.W.Grosse-Kunstleve,
V.B.Chen,
N.W.Moriarty,
D.C.Richardson,
J.S.Richardson,
and
P.D.Adams
(2012).
Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
381-390.
|
 |
|
|
|
|
 |
R.E.Hubbard
(2011).
Structure-based drug discovery and protein targets in the CNS.
|
| |
Neuropharmacology,
60,
7.
|
 |
|
|
|
|
 |
I.B.Müller,
F.Wu,
B.Bergmann,
J.Knöckel,
R.D.Walter,
H.Gehring,
and
C.Wrenger
(2009).
Poisoning pyridoxal 5-phosphate-dependent enzymes: a new strategy to target the malaria parasite Plasmodium falciparum.
|
| |
PLoS ONE,
4,
e4406.
|
 |
|
|
|
|
 |
Y.G.Kim,
S.Lee,
O.S.Kwon,
S.Y.Park,
S.J.Lee,
B.J.Park,
and
K.J.Kim
(2009).
Redox-switch modulation of human SSADH by dynamic catalytic loop.
|
| |
EMBO J,
28,
959-968.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.B.Berkowitz,
K.R.Karukurichi,
R.de la Salud-Bea,
D.L.Nelson,
and
C.D.McCune
(2008).
Use of Fluorinated Functionality in Enzyme Inhibitor Development: Mechanistic and Analytical Advantages.
|
| |
J Fluor Chem,
129,
731-742.
|
 |
|
|
|
|
 |
M.D.Clift,
and
R.B.Silverman
(2008).
Synthesis and evaluation of novel aromatic substrates and competitive inhibitors of GABA aminotransferase.
|
| |
Bioorg Med Chem Lett,
18,
3122-3125.
|
 |
|
|
|
|
 |
R.Meguro,
Y.Asano,
S.Odagiri,
C.Li,
and
K.Shoumura
(2008).
Cellular and subcellular localizations of nonheme ferric and ferrous iron in the rat brain: a light and electron microscopic study by the perfusion-Perls and -Turnbull methods.
|
| |
Arch Histol Cytol,
71,
205-222.
|
 |
|
|
|
|
 |
V.Rajaram,
P.Ratna Prasuna,
H.S.Savithri,
and
M.R.Murthy
(2008).
Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding.
|
| |
Proteins,
70,
429-441.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.R.Karukurichi,
R.de la Salud-Bea,
W.J.Jahng,
and
D.B.Berkowitz
(2007).
Examination of the new alpha-(2'Z-fluoro)vinyl trigger with lysine decarboxylase: the absolute stereochemistry dictates the reaction course.
|
| |
J Am Chem Soc,
129,
258-259.
|
 |
|
|
|
|
 |
M.D.Clift,
H.Ji,
G.P.Deniau,
D.O'Hagan,
and
R.B.Silverman
(2007).
Enantiomers of 4-amino-3-fluorobutanoic acid as substrates for gamma-aminobutyric acid aminotransferase. Conformational probes for GABA binding.
|
| |
Biochemistry,
46,
13819-13828.
|
 |
|
|
|
|
 |
Q.Wu,
Y.N.Liu,
H.Chen,
E.J.Molitor,
and
H.W.Liu
(2007).
A retro-evolution study of CDP-6-deoxy-D-glycero-L-threo-4-hexulose-3-dehydrase (E1) from Yersinia pseudotuberculosis: implications for C-3 deoxygenation in the biosynthesis of 3,6-dideoxyhexoses.
|
| |
Biochemistry,
46,
3759-3767.
|
 |
|
|
|
|
 |
D.B.Berkowitz,
B.Wu,
and
H.Li
(2006).
A formal [3,3]-sigmatropic rearrangement route to quaternary alpha-vinyl amino acids: use of allylic N-PMP trifluoroacetimidates.
|
| |
Org Lett,
8,
971-974.
|
 |
|
|
|
|
 |
S.M.Tripathi,
and
R.Ramachandran
(2006).
Overexpression, purification and crystallization of lysine epsilon-aminotransferase (Rv3290c) from Mycobacterium tuberculosis H37Rv.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
572-575.
|
 |
|
|
|
|
 |
V.Rajaram,
K.Prasad,
P.Ratna Prasuna,
N.Ramachandra,
S.R.Bharath,
H.S.Savithri,
and
M.R.Murthy
(2006).
Cloning, purification, crystallization and preliminary X-ray crystallographic analysis of the biosynthetic N-acetylornithine aminotransferases from Salmonella typhimurium and Escherichia coli.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
980-983.
|
 |
|
|
|
|
 |
R.O.Beleboni,
R.O.Carolino,
A.B.Pizzo,
L.Castellan-Baldan,
J.Coutinho-Netto,
W.F.dos Santos,
and
N.C.Coimbra
(2004).
Pharmacological and biochemical aspects of GABAergic neurotransmission: pathological and neuropsychobiological relationships.
|
| |
Cell Mol Neurobiol,
24,
707-728.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

