 |
PDBsum entry 1ng3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ng3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.19
- glycine oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glycine + O2 + H2O = glyoxylate + H2O2 + NH4+
|
 |
 |
 |
 |
 |
glycine
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
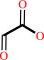 glyoxylate
glyoxylate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
42:2971-2981
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and mechanistic studies on ThiO, a glycine oxidase essential for thiamin biosynthesis in Bacillus subtilis.
|
|
E.C.Settembre,
P.C.Dorrestein,
J.H.Park,
A.M.Augustine,
T.P.Begley,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The thiO gene of Bacillus subtilis encodes an FAD-dependent glycine oxidase.
This enzyme is a homotetramer with a monomer molecular mass of 42 kDa. In this
paper, we demonstrate that ThiO is required for the biosynthesis of the thiazole
moiety of thiamin pyrophosphate and describe the structure of the enzyme with
N-acetylglycine bound at the active site. The closest structural relatives of
ThiO are sarcosine oxidase and d-amino acid oxidase. The ThiO structure, as well
as the observation that N-cyclopropylglycine is a good substrate, supports a
hydride transfer mechanism for the enzyme. A mechanistic proposal for the role
of ThiO in thiazole biosynthesis is also described.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.R.Challand,
F.T.Martins,
and
P.L.Roach
(2010).
Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli.
|
| |
J Biol Chem,
285,
5240-5248.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2010).
Oxidation of amines by flavoproteins.
|
| |
Arch Biochem Biophys,
493,
13-25.
|
 |
|
|
|
|
 |
A.Hazra,
A.Chatterjee,
and
T.P.Begley
(2009).
Biosynthesis of the thiamin thiazole in Bacillus subtilis: identification of the product of the thiazole synthase-catalyzed reaction.
|
| |
J Am Chem Soc,
131,
3225-3229.
|
 |
|
|
|
|
 |
C.T.Jurgenson,
T.P.Begley,
and
S.E.Ealick
(2009).
The structural and biochemical foundations of thiamin biosynthesis.
|
| |
Annu Rev Biochem,
78,
569-603.
|
 |
|
|
|
|
 |
H.Jørgensen,
K.F.Degnes,
H.Sletta,
E.Fjaervik,
A.Dikiy,
L.Herfindal,
P.Bruheim,
G.Klinkenberg,
H.Bredholt,
G.Nygård,
S.O.Døskeland,
T.E.Ellingsen,
and
S.B.Zotchev
(2009).
Biosynthesis of macrolactam BE-14106 involves two distinct PKS systems and amino acid processing enzymes for generation of the aminoacyl starter unit.
|
| |
Chem Biol,
16,
1109-1121.
|
 |
|
|
|
|
 |
M.Pedotti,
E.Rosini,
G.Molla,
T.Moschetti,
C.Savino,
B.Vallone,
and
L.Pollegioni
(2009).
Glyphosate resistance by engineering the flavoenzyme glycine oxidase.
|
| |
J Biol Chem,
284,
36415-36423.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Winkler,
A.Lyskowski,
S.Riedl,
M.Puhl,
T.M.Kutchan,
P.Macheroux,
and
K.Gruber
(2008).
A concerted mechanism for berberine bridge enzyme.
|
| |
Nat Chem Biol,
4,
739-741.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Martínez-Martínez,
J.Navarro-Fernández,
F.García-Carmona,
H.Takami,
and
A.Sánchez-Ferrer
(2008).
Characterization and structural modeling of a novel thermostable glycine oxidase from Geobacillus kaustophilus HTA426.
|
| |
Proteins,
70,
1429-1441.
|
 |
|
|
|
|
 |
L.Pollegioni,
G.Molla,
S.Sacchi,
E.Rosini,
R.Verga,
and
M.S.Pilone
(2008).
Properties and applications of microbial D: -amino acid oxidases: current state and perspectives.
|
| |
Appl Microbiol Biotechnol,
78,
1.
|
 |
|
|
|
|
 |
C.J.Carrell,
R.C.Bruckner,
D.Venci,
G.Zhao,
M.S.Jorns,
and
F.S.Mathews
(2007).
NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution.
|
| |
Structure,
15,
928-941.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.C.Ralph,
J.S.Hirschi,
M.A.Anderson,
W.W.Cleland,
D.A.Singleton,
and
P.F.Fitzpatrick
(2007).
Insights into the mechanism of flavoprotein-catalyzed amine oxidation from nitrogen isotope effects on the reaction of N-methyltryptophan oxidase.
|
| |
Biochemistry,
46,
7655-7664.
|
 |
|
|
|
|
 |
I.Martínez-Martínez,
C.Kaiser,
A.Rohde,
A.Ellert,
F.García-Carmona,
A.Sanchez-Ferrer,
and
R.Luttmann
(2007).
High-level production of bacillus subtilis glycine oxidase by fed-batch cultivation of recombinant Escherichia coli Rosetta (DE3).
|
| |
Biotechnol Prog,
23,
645-651.
|
 |
|
|
|
|
 |
M.Kriek,
F.Martins,
R.Leonardi,
S.A.Fairhurst,
D.J.Lowe,
and
P.L.Roach
(2007).
Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro.
|
| |
J Biol Chem,
282,
17413-17423.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2007).
Insights into the mechanisms of flavoprotein oxidases from kinetic isotope effects.
|
| |
J Labelled Comp Radiopharm,
50,
1016-1025.
|
 |
|
|
|
|
 |
A.Chatterjee,
X.Han,
F.W.McLafferty,
and
T.P.Begley
(2006).
Biosynthesis of thiamin thiazole: determination of the regiochemistry of the S/O acyl shift by using 1,4-dideoxy-D-xylulose-5-phosphate.
|
| |
Angew Chem Int Ed Engl,
45,
3507-3510.
|
 |
|
|
|
|
 |
C.Lehmann,
T.P.Begley,
and
S.E.Ealick
(2006).
Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis.
|
| |
Biochemistry,
45,
11-19.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.M.Downs
(2006).
Understanding microbial metabolism.
|
| |
Annu Rev Microbiol,
60,
533-559.
|
 |
|
|
|
|
 |
E.C.Ralph,
M.A.Anderson,
W.W.Cleland,
and
P.F.Fitzpatrick
(2006).
Mechanistic studies of the flavoenzyme tryptophan 2-monooxygenase: deuterium and 15N kinetic isotope effects on alanine oxidation by an L-amino acid oxidase.
|
| |
Biochemistry,
45,
15844-15852.
|
 |
|
|
|
|
 |
P.H.Godoi,
R.S.Galhardo,
D.D.Luche,
M.A.Van Sluys,
C.F.Menck,
and
G.Oliva
(2006).
Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana.
|
| |
J Biol Chem,
281,
30957-30966.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.P.Begley
(2006).
Cofactor biosynthesis: an organic chemist's treasure trove.
|
| |
Nat Prod Rep,
23,
15-25.
|
 |
|
|
|
|
 |
E.C.Ralph,
and
P.F.Fitzpatrick
(2005).
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase.
|
| |
Biochemistry,
44,
3074-3081.
|
 |
|
|
|
|
 |
V.I.Tishkov,
and
S.V.Khoronenkova
(2005).
D-Amino acid oxidase: structure, catalytic mechanism, and practical application.
|
| |
Biochemistry (Mosc),
70,
40-54.
|
 |
|
|
|
|
 |
J.M.Bujnicki,
Y.Oudjama,
M.Roovers,
S.Owczarek,
J.Caillet,
and
L.Droogmans
(2004).
Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA.
|
| |
RNA,
10,
1236-1242.
|
 |
|
|
|
|
 |
M.Mörtl,
K.Diederichs,
W.Welte,
G.Molla,
L.Motteran,
G.Andriolo,
M.S.Pilone,
and
L.Pollegioni
(2004).
Structure-function correlation in glycine oxidase from Bacillus subtilis.
|
| |
J Biol Chem,
279,
29718-29727.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.C.Martinez-Gomez,
M.Robers,
and
D.M.Downs
(2004).
Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily.
|
| |
J Biol Chem,
279,
40505-40510.
|
 |
|
|
|
|
 |
P.C.Dorrestein,
H.Zhai,
F.W.McLafferty,
and
T.P.Begley
(2004).
The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS.
|
| |
Chem Biol,
11,
1373-1381.
|
 |
|
|
|
|
 |
R.Leonardi,
and
P.L.Roach
(2004).
Thiamine biosynthesis in Escherichia coli: in vitro reconstitution of the thiazole synthase activity.
|
| |
J Biol Chem,
279,
17054-17062.
|
 |
|
|
|
|
 |
E.Settembre,
T.P.Begley,
and
S.E.Ealick
(2003).
Structural biology of enzymes of the thiamin biosynthesis pathway.
|
| |
Curr Opin Struct Biol,
13,
739-747.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

