 |
PDBsum entry 1ljl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ljl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Wild type pi258 s. Aureus arsenate reductase
|
|
Structure:
|
 |
Arsenate reductase. Chain: a. Synonym: arsenical pump modifier. Engineered: yes. Other_details: reduced
|
|
Source:
|
 |
Staphylococcus aureus. Organism_taxid: 1280. Gene: arsc. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.01Å
|
R-factor:
|
0.214
|
R-free:
|
0.259
|
|
|
Authors:
|
 |
J.Messens,J.C.Martins,K.Van Belle,E.Brosens,A.Desmyter,M.De Gieter, J.M.Wieruszeski,R.Willem,L.Wyns,I.Zegers
|
Key ref:

|
 |
J.Messens
et al.
(2002).
All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc Natl Acad Sci U S A,
99,
8506-8511.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
21-Apr-02
|
Release date:
|
07-Aug-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0A006
(ARSC_STAAU) -
Arsenate reductase from Staphylococcus aureus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
131 a.a.
130 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.20.4.4
- arsenate reductase (thioredoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
arsenate + [thioredoxin]-dithiol + H+ = arsenite + [thioredoxin]- disulfide + H2O
|
 |
 |
 |
 |
 |
 arsenate
arsenate
|
+
|
[thioredoxin]-dithiol
|
+
|
H(+)
|
=
|
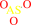 arsenite
arsenite
|
+
|
[thioredoxin]- disulfide
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
99:8506-8511
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
|
|
J.Messens,
J.C.Martins,
K.Van Belle,
E.Brosens,
A.Desmyter,
M.De Gieter,
J.M.Wieruszeski,
R.Willem,
L.Wyns,
I.Zegers.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The mechanism of pI258 arsenate reductase (ArsC) catalyzed arsenate reduction,
involving its P-loop structural motif and three redox active cysteines, has been
unraveled. All essential intermediates are visualized with x-ray
crystallography, and NMR is used to map dynamic regions in a key disulfide
intermediate. Steady-state kinetics of ArsC mutants gives a view of the crucial
residues for catalysis. ArsC combines a phosphatase-like nucleophilic
displacement reaction with a unique intramolecular disulfide bond cascade.
Within this cascade, the formation of a disulfide bond triggers a reversible
"conformational switch" that transfers the oxidative equivalents to
the surface of the protein, while releasing the reduced substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. (A) Scheme of the reaction mechanism of pI258
ArsC. (1) The nucleophilic attack of the thiol of Cys-10; (2)
the formation of a covalent Cys-10-HAsO[  -
]intermediate; (3) the nucleophilic attack of the thiol of
Cys-82 with arsenite release; (4) the formation of a
Cys-10-Cys-82 intermediate and the nucleophilic attack of the
thiol of Cys-89; (5) the formation of a Cys-82-Cys-89 disulfide.
(B-F) A stereo view of the 2F[o] -
]intermediate; (3) the nucleophilic attack of the thiol of
Cys-82 with arsenite release; (4) the formation of a
Cys-10-Cys-82 intermediate and the nucleophilic attack of the
thiol of Cys-89; (5) the formation of a Cys-82-Cys-89 disulfide.
(B-F) A stereo view of the 2F[o]  F[c]
electron density maps contoured at 1.0 F[c]
electron density maps contoured at 1.0  placed
next to its corresponding reaction step in A. (B) The P-loop
(residues 10-17) in the structure of reduced wild-type ArsC with
Cys-10 in the center of the image. The P-loop is fully
structured, despite the absence of bound oxyanion (2.0 Å).
(C) In the structure of C15A ArsC-HAsO[ placed
next to its corresponding reaction step in A. (B) The P-loop
(residues 10-17) in the structure of reduced wild-type ArsC with
Cys-10 in the center of the image. The P-loop is fully
structured, despite the absence of bound oxyanion (2.0 Å).
(C) In the structure of C15A ArsC-HAsO[  -
], an arsenic is covalently bound on Cys-10, surrounded by three
oxygens in a plane and a water molecule opposite the sulfur of
Cys-10 (1.4 Å). (D) Oxidized ArsC C89L with the
intermediate Cys-10-Cys-82 disulfide bond (1.6 Å). (E) A
view on the flexible looped-out region of oxidized ArsC C89L,
where Cys-89 has left the hydrophobic core and is replaced by
Leu-92 upon Cys-10-Cys-82 formation. The electron density in
this highly flexible region is not so well defined. (F) A view
on the surface of oxidized ArsC C10SC15A (6) with the
Cys-82-Cys-89 disulfide bond. -
], an arsenic is covalently bound on Cys-10, surrounded by three
oxygens in a plane and a water molecule opposite the sulfur of
Cys-10 (1.4 Å). (D) Oxidized ArsC C89L with the
intermediate Cys-10-Cys-82 disulfide bond (1.6 Å). (E) A
view on the flexible looped-out region of oxidized ArsC C89L,
where Cys-89 has left the hydrophobic core and is replaced by
Leu-92 upon Cys-10-Cys-82 formation. The electron density in
this highly flexible region is not so well defined. (F) A view
on the surface of oxidized ArsC C10SC15A (6) with the
Cys-82-Cys-89 disulfide bond.
|
 |
Figure 4.
Fig. 4. The movement of the "conformational switch" in
the flexible segment (residues 80-98) trapped in four different
ArsC crystals. Starting with a helix in the reduced ArsC wild
type (blue), via oxidized ArsC C89L (Cys-10-Cys-82) in the first
(yellow) and in the second (red) molecule in the asymmetric unit
to finally looping out to form the C82-C89 disulfide (green).
The two arrows indicate the movement of L92 and C89.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Yu,
B.Xia,
and
C.Jin
(2011).
(1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803.
|
| |
Biomol NMR Assign,
5,
85-87.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Jain,
B.Saluja,
A.Gupta,
S.S.Marla,
and
R.Goel
(2011).
Validation of Arsenic Resistance in Bacillus cereus Strain AG27 by Comparative Protein Modeling of arsC Gene Product.
|
| |
Protein J,
30,
91.
|
 |
|
|
|
|
 |
E.Pedone,
D.Limauro,
K.D'Ambrosio,
G.De Simone,
and
S.Bartolucci
(2010).
Multiple catalytically active thioredoxin folds: a winning strategy for many functions.
|
| |
Cell Mol Life Sci,
67,
3797-3814.
|
 |
|
|
|
|
 |
M.A.Wouters,
S.W.Fan,
and
N.L.Haworth
(2010).
Disulfides as redox switches: from molecular mechanisms to functional significance.
|
| |
Antioxid Redox Signal,
12,
53-91.
|
 |
|
|
|
|
 |
E.Ordóñez,
K.Van Belle,
G.Roos,
S.De Galan,
M.Letek,
J.A.Gil,
L.Wyns,
L.M.Mateos,
and
J.Messens
(2009).
Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange.
|
| |
J Biol Chem,
284,
15107-15116.
|
 |
|
|
|
|
 |
G.Roos,
N.Foloppe,
K.Van Laer,
L.Wyns,
L.Nilsson,
P.Geerlings,
and
J.Messens
(2009).
How thioredoxin dissociates its mixed disulfide.
|
| |
PLoS Comput Biol,
5,
e1000461.
|
 |
|
|
|
|
 |
L.López-Maury,
A.M.Sánchez-Riego,
J.C.Reyes,
and
F.J.Florencio
(2009).
The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803.
|
| |
J Bacteriol,
191,
3534-3543.
|
 |
|
|
|
|
 |
S.M.Marino,
and
V.N.Gladyshev
(2009).
A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues.
|
| |
PLoS Comput Biol,
5,
e1000383.
|
 |
|
|
|
|
 |
S.W.Fan,
R.A.George,
N.L.Haworth,
L.L.Feng,
J.Y.Liu,
and
M.A.Wouters
(2009).
Conformational changes in redox pairs of protein structures.
|
| |
Protein Sci,
18,
1745-1765.
|
 |
|
|
|
|
 |
D.S.Touw,
C.E.Nordman,
J.A.Stuckey,
and
V.L.Pecoraro
(2007).
Identifying important structural characteristics of arsenic resistance proteins by using designed three-stranded coiled coils.
|
| |
Proc Natl Acad Sci U S A,
104,
11969-11974.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.De la Rosa,
J.G.Parsons,
A.Martinez-Martinez,
J.R.Peralta-Videa,
and
J.L.Gardea-Torresdey
(2006).
Spectroscopic study of the impact of arsenic speciation on arsenic/phosphorus uptake and plant growth in tumbleweed (Salsola kali).
|
| |
Environ Sci Technol,
40,
1991-1996.
|
 |
|
|
|
|
 |
G.Roos,
S.Loverix,
E.Brosens,
K.Van Belle,
L.Wyns,
P.Geerlings,
and
J.Messens
(2006).
The activation of electrophile, nucleophile and leaving group during the reaction catalysed by pI258 arsenate reductase.
|
| |
Chembiochem,
7,
981-989.
|
 |
|
|
|
|
 |
A.Salmeen,
and
D.Barford
(2005).
Functions and mechanisms of redox regulation of cysteine-based phosphatases.
|
| |
Antioxid Redox Signal,
7,
560-577.
|
 |
|
|
|
|
 |
F.S.Islam,
R.L.Pederick,
A.G.Gault,
L.K.Adams,
D.A.Polya,
J.M.Charnock,
and
J.R.Lloyd
(2005).
Interactions between the Fe(III)-reducing bacterium Geobacter sulfurreducens and arsenate, and capture of the metalloid by biogenic Fe(II).
|
| |
Appl Environ Microbiol,
71,
8642-8648.
|
 |
|
|
|
|
 |
S.Silver,
and
L.T.Phung
(2005).
Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic.
|
| |
Appl Environ Microbiol,
71,
599-608.
|
 |
|
|
|
|
 |
J.Messens,
I.Van Molle,
P.Vanhaesebrouck,
K.Van Belle,
K.Wahni,
J.C.Martins,
L.Wyns,
and
R.Loris
(2004).
The structure of a triple mutant of pI258 arsenate reductase from Staphylococcus aureus and its 5-thio-2-nitrobenzoic acid adduct.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1180-1184.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.I.Leichert,
C.Scharf,
and
M.Hecker
(2003).
Global characterization of disulfide stress in Bacillus subtilis.
|
| |
J Bacteriol,
185,
1967-1975.
|
 |
|
|
|
|
 |
L.López-Maury,
F.J.Florencio,
and
J.C.Reyes
(2003).
Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803.
|
| |
J Bacteriol,
185,
5363-5371.
|
 |
|
|
|
|
 |
P.Retailleau,
and
T.Prangé
(2003).
Phasing power at the K absorption edge of organic arsenic.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
887-896.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Li,
J.D.Haile,
and
P.J.Kennelly
(2003).
An arsenate reductase from Synechocystis sp. strain PCC 6803 exhibits a novel combination of catalytic characteristics.
|
| |
J Bacteriol,
185,
6780-6789.
|
 |
|
|
|
|
 |
S.Silver
(2003).
Bacterial silver resistance: molecular biology and uses and misuses of silver compounds.
|
| |
FEMS Microbiol Rev,
27,
341-353.
|
 |
|
|
|
|
 |
R.Mukhopadhyay,
B.P.Rosen,
L.T.Phung,
and
S.Silver
(2002).
Microbial arsenic: from geocycles to genes and enzymes.
|
| |
FEMS Microbiol Rev,
26,
311-325.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

