 |
PDBsum entry 1js3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.28
- aromatic-L-amino-acid decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Dopa Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-dopa + H+ = dopamine + CO2
|
|
2.
|
5-hydroxy-L-tryptophan + H+ = serotonin + CO2
|
|
 |
 |
 |
 |
 |
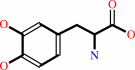 L-dopa
L-dopa
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
=
|
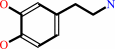 dopamine
dopamine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
5-hydroxy-L-tryptophan
Bound ligand (Het Group name = )
matches with 68.42% similarity
|
+
|
H(+)
|
=
|
 serotonin
serotonin
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
8:963-967
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
|
|
P.Burkhard,
P.Dominici,
C.Borri-Voltattorni,
J.N.Jansonius,
V.N.Malashkevich.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
DOPA decarboxylase (DDC) is responsible for the synthesis of the key
neurotransmitters dopamine and serotonin via decarboxylation of
L-3,4-dihydroxyphenylalanine (L-DOPA) and L-5-hydroxytryptophan, respectively.
DDC has been implicated in a number of clinic disorders, including Parkinson's
disease and hypertension. Peripheral inhibitors of DDC are currently used to
treat these diseases. We present the crystal structures of ligand-free DDC and
its complex with the anti-Parkinson drug carbiDOPA. The inhibitor is bound to
the enzyme by forming a hydrazone linkage with the cofactor, and its catechol
ring is deeply buried in the active site cleft. The structures provide the
molecular basis for the development of new inhibitors of DDC with better
pharmacological characteristics.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Stereo view ribbon diagram of the polypeptide backbone
of DDC. The view is directly down the two-fold symmetry axis.
One monomer is completely red, whereas the other is green
(N-terminal domain), cyan (large domain) and blue (small
domain). The cofactors (PLP) and the inhibitors (carbiDOPA) are
shown in ball-and-stick representation in yellow. The N-terminal
domain of one monomer packs on top of the other monomer,
resulting in an extended dimer interface. The picture was drawn
with MOLSCRIPT50 and RASTER3D^51.
|
 |
Figure 3.
Figure 3. Active site cleft of DDC in complex with carbiDOPA.
a, Stereo view of the electron density of the inhibitor
carbiDOPA. The difference electron density (|F[o]| - |F[c]| map
with the inhibitor excluded from the phase calculation) in red,
contoured at 4  ,
is superimposed onto the inhibitor model. Nitrogen, phosphate
and oxygen atoms are marked blue, cyan and red, respectively.
Carbon atoms are colored in yellow for the enzyme, in magenta
for the PLP -carbiDOPA complex and in orange for the residues of
the other monomer. Hydrogen bonds are indicated in green dotted
lines. b, Detailed view of the hydrogen bond interactions,
including all structural water molecules in the active site.
Color code as in (a). c, A model of modified carbiDOPA with an
additional 2' hydroxyl group (cyan). The newly established
hydrogen bonds to the structural water molecules and the
hydrazone nitrogen are indicated as dotted lines (cyan).
Otherwise, the color code is as in (a). The picture was drawn
with MOLSCRIPT50 and RASTER3D^51. ,
is superimposed onto the inhibitor model. Nitrogen, phosphate
and oxygen atoms are marked blue, cyan and red, respectively.
Carbon atoms are colored in yellow for the enzyme, in magenta
for the PLP -carbiDOPA complex and in orange for the residues of
the other monomer. Hydrogen bonds are indicated in green dotted
lines. b, Detailed view of the hydrogen bond interactions,
including all structural water molecules in the active site.
Color code as in (a). c, A model of modified carbiDOPA with an
additional 2' hydroxyl group (cyan). The newly established
hydrogen bonds to the structural water molecules and the
hydrazone nitrogen are indicated as dotted lines (cyan).
Otherwise, the color code is as in (a). The picture was drawn
with MOLSCRIPT50 and RASTER3D^51.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2001,
8,
963-967)
copyright 2001.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.E.Hubbard
(2011).
Structure-based drug discovery and protein targets in the CNS.
|
| |
Neuropharmacology,
60,
7.
|
 |
|
|
|
|
 |
Q.Han,
H.Ding,
H.Robinson,
B.M.Christensen,
and
J.Li
(2010).
Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase.
|
| |
PLoS One,
5,
e8826.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Zhang,
J.Y.Zhou,
M.H.Chin,
A.A.Schepmoes,
V.A.Petyuk,
K.K.Weitz,
B.O.Petritis,
M.E.Monroe,
D.G.Camp,
S.A.Wood,
W.P.Melega,
D.J.Bigelow,
D.J.Smith,
W.J.Qian,
and
R.D.Smith
(2010).
Region-specific protein abundance changes in the brain of MPTP-induced Parkinson's disease mouse model.
|
| |
J Proteome Res,
9,
1496-1509.
|
 |
|
|
|
|
 |
Y.L.Lin,
and
J.Gao
(2010).
Internal proton transfer in the external pyridoxal 5'-phosphate Schiff base in dopa decarboxylase.
|
| |
Biochemistry,
49,
84-94.
|
 |
|
|
|
|
 |
K.E.Fujimori
(2009).
Characterization of the Regulatory Region of the Dopa Decarboxylase Gene in Medaka: An in vivo Green Fluorescent Protein Reporter Assay Combined with a Simple TA-Cloning Method.
|
| |
Mol Biotechnol,
41,
224-235.
|
 |
|
|
|
|
 |
L.R.Hofto,
C.E.Lee,
and
M.Cafiero
(2009).
The importance of aromatic-type interactions in serotonin synthesis: protein-ligand interactions in tryptophan hydroxylase and aromatic amino acid decarboxylase.
|
| |
J Comput Chem,
30,
1111-1115.
|
 |
|
|
|
|
 |
M.Wiltgen,
and
G.P.Tilz
(2009).
Homology modelling: a review about the method on hand of the diabetic antigen GAD 65 structure prediction.
|
| |
Wien Med Wochenschr,
159,
112-125.
|
 |
|
|
|
|
 |
A.A.Moya-García,
J.Ruiz-Pernía,
S.Martí,
F.Sánchez-Jiménez,
and
I.Tuñón
(2008).
Analysis of the decarboxylation step in mammalian histidine decarboxylase. A computational study.
|
| |
J Biol Chem,
283,
12393-12401.
|
 |
|
|
|
|
 |
D.Mukhopadhyay,
K.S.Howell,
H.Riezman,
and
G.Capitani
(2008).
Identifying key residues of sphinganine-1-phosphate lyase for function in vivo and in vitro.
|
| |
J Biol Chem,
283,
20159-20169.
|
 |
|
|
|
|
 |
G.Fenalti,
R.H.Law,
A.M.Buckle,
C.Langendorf,
K.Tuck,
C.J.Rosado,
N.G.Faux,
K.Mahmood,
C.S.Hampe,
J.P.Banga,
M.Wilce,
J.Schmidberger,
J.Rossjohn,
O.El-Kabbani,
R.N.Pike,
A.I.Smith,
I.R.Mackay,
M.J.Rowley,
and
J.C.Whisstock
(2007).
GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
|
| |
Nat Struct Mol Biol,
14,
280-286.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.E.Ippolito,
M.E.Merritt,
F.Bäckhed,
K.L.Moulder,
S.Mennerick,
J.K.Manchester,
S.T.Gammon,
D.Piwnica-Worms,
and
J.I.Gordon
(2006).
Linkage between cellular communications, energy utilization, and proliferation in metastatic neuroendocrine cancers.
|
| |
Proc Natl Acad Sci U S A,
103,
12505-12510.
|
 |
|
|
|
|
 |
J.Stetefeld,
M.Jenny,
and
P.Burkhard
(2006).
Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
|
| |
Proc Natl Acad Sci U S A,
103,
13688-13693.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Gütschow,
and
M.Meusel
(2006).
[Enzyme inhibitors in Parkinson treatment]
|
| |
Pharm Unserer Zeit,
35,
218-225.
|
 |
|
|
|
|
 |
A.A.Moya-Garcia,
M.A.Medina,
and
F.Sánchez-Jiménez
(2005).
Mammalian histidine decarboxylase: from structure to function.
|
| |
Bioessays,
27,
57-63.
|
 |
|
|
|
|
 |
G.Capitani,
D.De Biase,
H.Gut,
S.Ahmed,
and
M.G.Grütter
(2005).
Structural model of human GAD65: prediction and interpretation of biochemical and immunogenic features.
|
| |
Proteins,
59,
7.
|
 |
|
|
|
|
 |
J.Wei,
and
J.Y.Wu
(2005).
Structural and functional analysis of cysteine residues in human glutamate decarboxylase 65 (GAD65) and GAD67.
|
| |
J Neurochem,
93,
624-633.
|
 |
|
|
|
|
 |
M.A.Medina,
F.Correa-Fiz,
C.Rodríguez-Caso,
and
F.Sánchez-Jiménez
(2005).
A comprehensive view of polyamine and histamine metabolism to the light of new technologies.
|
| |
J Cell Mol Med,
9,
854-864.
|
 |
|
|
|
|
 |
M.J.Alkema,
M.Hunter-Ensor,
N.Ringstad,
and
H.R.Horvitz
(2005).
Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system.
|
| |
Neuron,
46,
247-260.
|
 |
|
|
|
|
 |
T.Nakai,
N.Nakagawa,
N.Maoka,
R.Masui,
S.Kuramitsu,
and
N.Kamiya
(2005).
Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia.
|
| |
EMBO J,
24,
1523-1536.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Paiardini,
F.Bossa,
and
S.Pascarella
(2004).
Evolutionarily conserved regions and hydrophobic contacts at the superfamily level: The case of the fold-type I, pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Protein Sci,
13,
2992-3005.
|
 |
|
|
|
|
 |
E.E.Hare,
and
C.M.Loer
(2004).
Function and evolution of the serotonin-synthetic bas-1 gene and other aromatic amino acid decarboxylase genes in Caenorhabditis.
|
| |
BMC Evol Biol,
4,
24.
|
 |
|
|
|
|
 |
C.Rodríguez-Caso,
D.Rodríguez-Agudo,
A.A.Moya-García,
I.Fajardo,
M.A.Medina,
V.Subramaniam,
and
F.Sánchez-Jiménez
(2003).
Local changes in the catalytic site of mammalian histidine decarboxylase can affect its global conformation and stability.
|
| |
Eur J Biochem,
270,
4376-4387.
|
 |
|
|
|
|
 |
G.Capitani,
D.De Biase,
C.Aurizi,
H.Gut,
F.Bossa,
and
M.G.Grütter
(2003).
Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase.
|
| |
EMBO J,
22,
4027-4037.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.G.Cheong,
J.C.Escalante-Semerena,
and
I.Rayment
(2002).
Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
|
| |
Biochemistry,
41,
9079-9089.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bertoldi,
M.Gonsalvi,
R.Contestabile,
and
C.B.Voltattorni
(2002).
Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase.
|
| |
J Biol Chem,
277,
36357-36362.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

