
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of xanthine dehydrogenase from rhodobacter capsulatus
|
|
Structure:
|
 |
Xanthine dehydrogenase, chain a. Chain: a, c, e, g. Fragment: chain a, residues 1-462. Synonym: xd. Engineered: yes. Xanthine dehydrogenase, chain b. Chain: b, d, f, h. Fragment: chain b, residues 1-777. Synonym: xd.
|
|
Source:
|
 |
Rhodobacter capsulatus. Organism_taxid: 1061. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Hexamer (from
)
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.215
|
R-free:
|
0.252
|
|
|
Authors:
|
 |
J.J.Truglio,K.Theis,S.Leimkuhler,R.Rappa,K.V.Rajagopalan,C.Kisker
|
Key ref:

|
 |
J.J.Truglio
et al.
(2002).
Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus.
Structure,
10,
115-125.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-Aug-01
|
Release date:
|
11-Jan-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.1.1.1.204
- Transferred entry: 1.17.1.4.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
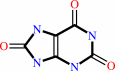
Xanthine + NAD+ + H2O = urate + NADH
|
 |
 |
 |
 |
 |
Bound ligand (Het Group name = )
matches with 45.00% similarity
|
+
|
Bound ligand (Het Group name = )
matches with 76.00% similarity
|
+
|
|
=
|
|
+
|

|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Molybdenum
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
10:115-125
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus.
|
|
J.J.Truglio,
K.Theis,
S.Leimkühler,
R.Rappa,
K.V.Rajagopalan,
C.Kisker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Xanthine dehydrogenase (XDH), a complex molybdo/iron-sulfur/flavoprotein,
catalyzes the oxidation of hypoxanthine to xanthine followed by oxidation of
xanthine to uric acid with concomitant reduction of NAD+. The 2.7 A resolution
structure of Rhodobacter capsulatus XDH reveals that the bacterial and bovine
XDH have highly similar folds despite differences in subunit composition. The
NAD+ binding pocket of the bacterial XDH resembles that of the dehydrogenase
form of the bovine enzyme rather than that of the oxidase form, which reduces
O(2) instead of NAD+. The drug allopurinol is used to treat XDH-catalyzed uric
acid build-up occurring in gout or during cancer chemotherapy. As a hypoxanthine
analog, it is oxidized to alloxanthine, which cannot be further oxidized but
acts as a tight binding inhibitor of XDH. The 3.0 A resolution structure of the
XDH-alloxanthine complex shows direct coordination of alloxanthine to the
molybdenum via a nitrogen atom. These results provide a starting point for the
rational design of new XDH inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Schematic Representation of Protein-Moco
InteractionsDashed lines indicate hydrogen bonds. In addition,
the aromatic side chain of Phe-B228 stacks with the pterin rings
(not shown).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2002,
10,
115-125)
copyright 2002.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Neumann,
and
S.Leimkühler
(2011).
The role of system-specific molecular chaperones in the maturation of molybdoenzymes in bacteria.
|
| |
Biochem Res Int,
2011,
850924.
|
 |
|
|
|
|
 |
M.C.Gomez-Cabrera,
G.L.Close,
A.Kayani,
A.McArdle,
J.Viña,
and
M.J.Jackson
(2010).
Effect of xanthine oxidase-generated extracellular superoxide on skeletal muscle force generation.
|
| |
Am J Physiol Regul Integr Comp Physiol,
298,
R2-R8.
|
 |
|
|
|
|
 |
Y.Fu,
R.Zhang,
D.Lu,
H.Liu,
K.Chandrashekar,
L.A.Juncos,
and
R.Liu
(2010).
NOX2 is the primary source of angiotensin II-induced superoxide in the macula densa.
|
| |
Am J Physiol Regul Integr Comp Physiol,
298,
R707-R712.
|
 |
|
|
|
|
 |
G.Schwarz,
R.R.Mendel,
and
M.W.Ribbe
(2009).
Molybdenum cofactors, enzymes and pathways.
|
| |
Nature,
460,
839-847.
|
 |
|
|
|
|
 |
J.M.Pauff,
H.Cao,
and
R.Hille
(2009).
Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE.
|
| |
J Biol Chem,
284,
8760-8767.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Pauff,
and
R.Hille
(2009).
Inhibition studies of bovine xanthine oxidase by luteolin, silibinin, quercetin, and curcumin.
|
| |
J Nat Prod,
72,
725-731.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
S.Schumann,
M.Terao,
E.Garattini,
M.Saggu,
F.Lendzian,
P.Hildebrandt,
and
S.Leimkühler
(2009).
Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1.
|
| |
PLoS ONE,
4,
e5348.
|
 |
|
|
|
|
 |
U.Dietzel,
J.Kuper,
J.A.Doebbler,
A.Schulte,
J.J.Truglio,
S.Leimkühler,
and
C.Kisker
(2009).
Mechanism of Substrate and Inhibitor Binding of Rhodobacter capsulatus Xanthine Dehydrogenase.
|
| |
J Biol Chem,
284,
8768-8776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Pauff,
J.Zhang,
C.E.Bell,
and
R.Hille
(2008).
Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine.
|
| |
J Biol Chem,
283,
4818-4824.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Li,
T.A.Müller,
B.A.Fraser,
and
R.P.Hausinger
(2008).
Characterization of active site variants of xanthine hydroxylase from Aspergillus nidulans.
|
| |
Arch Biochem Biophys,
470,
44-53.
|
 |
|
|
|
|
 |
S.Chaves,
M.Gil,
S.Canário,
R.Jelic,
M.J.Romão,
J.Trincão,
E.Herdtweck,
J.Sousa,
C.Diniz,
P.Fresco,
and
M.A.Santos
(2008).
Biologically relevant O,S-donor compounds. Synthesis, molybdenum complexation and xanthine oxidase inhibition.
|
| |
Dalton Trans,
(),
1773-1782.
|
 |
|
|
|
|
 |
S.Schumann,
M.Saggu,
N.Möller,
S.D.Anker,
F.Lendzian,
P.Hildebrandt,
and
S.Leimkühler
(2008).
The mechanism of assembly and cofactor insertion into Rhodobacter capsulatus xanthine dehydrogenase.
|
| |
J Biol Chem,
283,
16602-16611.
|
 |
|
|
|
|
 |
T.Nishino,
K.Okamoto,
B.T.Eger,
E.F.Pai,
and
T.Nishino
(2008).
Mammalian xanthine oxidoreductase - mechanism of transition from xanthine dehydrogenase to xanthine oxidase.
|
| |
FEBS J,
275,
3278-3289.
|
 |
|
|
|
|
 |
A.M.Burroughs,
S.Balaji,
L.M.Iyer,
and
L.Aravind
(2007).
Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold.
|
| |
Biol Direct,
2,
18.
|
 |
|
|
|
|
 |
A.Thapper,
D.R.Boer,
C.D.Brondino,
J.J.Moura,
and
M.J.Romão
(2007).
Correlating EPR and X-ray structural analysis of arsenite-inhibited forms of aldehyde oxidoreductase.
|
| |
J Biol Inorg Chem,
12,
353-366.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.M.Montero-Morán,
M.Li,
E.Rendòn-Huerta,
F.Jourdan,
D.J.Lowe,
A.W.Stumpff-Kane,
M.Feig,
C.Scazzocchio,
and
R.P.Hausinger
(2007).
Purification and characterization of the FeII- and alpha-ketoglutarate-dependent xanthine hydroxylase from Aspergillus nidulans.
|
| |
Biochemistry,
46,
5293-5304.
|
 |
|
|
|
|
 |
J.M.Pauff,
C.F.Hemann,
N.Jünemann,
S.Leimkühler,
and
R.Hille
(2007).
The role of arginine 310 in catalysis and substrate specificity in xanthine dehydrogenase from Rhodobacter capsulatus.
|
| |
J Biol Chem,
282,
12785-12790.
|
 |
|
|
|
|
 |
K.Pal,
P.K.Chaudhury,
and
S.Sarkar
(2007).
Structure of the Michaelis complex and function of the catalytic center in the reductive half-reaction of computational and synthetic models of sulfite oxidase.
|
| |
Chem Asian J,
2,
956-964.
|
 |
|
|
|
|
 |
C.D.Brondino,
M.J.Romão,
I.Moura,
and
J.J.Moura
(2006).
Molybdenum and tungsten enzymes: the xanthine oxidase family.
|
| |
Curr Opin Chem Biol,
10,
109-114.
|
 |
|
|
|
|
 |
M.Neumann,
M.Schulte,
N.Jünemann,
W.Stöcklein,
and
S.Leimkühler
(2006).
Rhodobacter capsulatus XdhC is involved in molybdenum cofactor binding and insertion into xanthine dehydrogenase.
|
| |
J Biol Chem,
281,
15701-15708.
|
 |
|
|
|
|
 |
P.Pacher,
A.Nivorozhkin,
and
C.Szabó
(2006).
Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol.
|
| |
Pharmacol Rev,
58,
87.
|
 |
|
|
|
|
 |
D.R.Boer,
A.Müller,
S.Fetzner,
D.J.Lowe,
and
M.J.Romão
(2005).
On the purification and preliminary crystallographic analysis of isoquinoline 1-oxidoreductase from Brevundimonas diminuta 7.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
137-140.
|
 |
|
|
|
|
 |
H.Cheng,
and
N.V.Grishin
(2005).
DOM-fold: a structure with crossing loops found in DmpA, ornithine acetyltransferase, and molybdenum cofactor-binding domain.
|
| |
Protein Sci,
14,
1902-1910.
|
 |
|
|
|
|
 |
U.Kappler,
and
S.Bailey
(2005).
Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit.
|
| |
J Biol Chem,
280,
24999-25007.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.E.Berry,
and
J.M.Hare
(2004).
Xanthine oxidoreductase and cardiovascular disease: molecular mechanisms and pathophysiological implications.
|
| |
J Physiol,
555,
589-606.
|
 |
|
|
|
|
 |
I.Bonin,
B.M.Martins,
V.Purvanov,
S.Fetzner,
R.Huber,
and
H.Dobbek
(2004).
Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
|
| |
Structure,
12,
1425-1435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.J.Moura,
C.D.Brondino,
J.Trincão,
and
M.J.Romão
(2004).
Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases.
|
| |
J Biol Inorg Chem,
9,
791-799.
|
 |
|
|
|
|
 |
L.Loschi,
S.J.Brokx,
T.L.Hills,
G.Zhang,
M.G.Bertero,
A.L.Lovering,
J.H.Weiner,
and
N.C.Strynadka
(2004).
Structural and biochemical identification of a novel bacterial oxidoreductase.
|
| |
J Biol Chem,
279,
50391-50400.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Unciuleac,
E.Warkentin,
C.C.Page,
M.Boll,
and
U.Ermler
(2004).
Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow.
|
| |
Structure,
12,
2249-2256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Leimkühler,
A.L.Stockert,
K.Igarashi,
T.Nishino,
and
R.Hille
(2004).
The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase.
|
| |
J Biol Chem,
279,
40437-40444.
|
 |
|
|
|
|
 |
K.Parschat,
B.Hauer,
R.Kappl,
R.Kraft,
J.Huttermann,
and
S.Fetzner
(2003).
Gene cluster of Arthrobacter ilicis Ru61a involved in the degradation of quinaldine to anthranilate: characterization and functional expression of the quinaldine 4-oxidase qoxLMS genes.
|
| |
J Biol Chem,
278,
27483-27494.
|
 |
|
|
|
|
 |
N.V.Ivanov,
F.Hubálek,
M.Trani,
and
D.E.Edmondson
(2003).
Factors involved in the assembly of a functional molybdopyranopterin center in recombinant Comamonas acidovorans xanthine dehydrogenase.
|
| |
Eur J Biochem,
270,
4744-4754.
|
 |
|
|
|
|
 |
S.Leimkuhler,
R.Hodson,
G.N.George,
and
K.V.Rajagopalan
(2003).
Recombinant Rhodobacter capsulatus xanthine dehydrogenase, a useful model system for the characterization of protein variants leading to xanthinuria I in humans.
|
| |
J Biol Chem,
278,
20802-20811.
|
 |
|
|
|
|
 |
R.Hille
(2002).
Molybdenum and tungsten in biology.
|
| |
Trends Biochem Sci,
27,
360-367.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

