 |
PDBsum entry 1j0a

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.99.7
- 1-aminocyclopropane-1-carboxylate deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-aminocyclopropane-1-carboxylate + H2O = 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
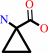 1-aminocyclopropane-1-carboxylate
1-aminocyclopropane-1-carboxylate
|
+
|
H2O
|
=
|
2-oxobutanoate
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
341:999
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii.
|
|
A.Fujino,
T.Ose,
M.Yao,
T.Tokiwano,
M.Honma,
N.Watanabe,
I.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
1-Aminocyclopropane-l-carboxylate deaminase (ACCD) is a pyridoxal 5/-phosphate
dependent enzyme that shows deaminase activity toward ACC, a precursor of plant
hormone ethylene. ACCD from some soil bacteria has been reported to be able to
break the cyclopropane ring of ACC to yield a-ketobutyrate and ammonia. We
reported the crystal structure of ACCD from the yeast Hansenula saturnus in the
absence/presence of substrate ACC, and proposed its ingenious reaction
mechanisms. In order to study the enzyme further, we overexpressed the ACCD
homologue protein (phAHP) from the fully decoded hyperthermophilic archearon,
Pyrococcus horikoshii OT3. However, phAHP does not show ACCD activity at high
temperature as well as at room temperature, though it has significant sequence
similarity. Instead of ACCD activity, the GC-MS analysis and enzymatic method
show that phAHP has deaminase activity toward L and D-serine. Here, we present
the crystal structures of the native and ACC-complexed phAHP. The overall
topology of the phAHP structure is very similar to that of ACCD; however,
critical differences were observed around the active site. Here, the differences
of enzymatic activity between phAHP and ACCD are discussed based on the
structural differences of these two proteins. We suggest that the catalytic
disagreement between these two enzymes comes from the difference of the residues
near the pyridine ring of pyridoxal 5'-phosphate (PLP), not the difference of
the catalytic residues themselves. We also propose a condition necessary in the
primary sequence to have ACCD activity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Ribbon representation of the two dimers. The
crystallographic dimer (AA dimer) is shown in red, the
non-crystallographic (NCS) dimer (BC dimer) in blue and green.
The PLP is shown in yellow using the ball-and-stick model. The
Figure is a view from the NCS 2-fold axis.
|
 |
Figure 10.
Figure 10. Comparison of active sites between phAHP and
hACCD. (a) Superimposition of ACC complexed phAHP (yellow) and
ACC complexed hACCD (K51T, green). (b) Stereo view showing the
active site of phAHP. The main chain is colored blue and the
side-chains that form the pathway from the solvent region are
colored gray. PLP is shown in yellow ball-and-stick model. The
black arrow indicates the plausible substrate pathway. (c)
Stereo view showing the active site of hACCD. The main-chain is
colored green and the side-chains that form the cavity from the
solvent region are colored gray. PLP is shown in yellow
ball-and-stick model. The orientation is the same as that in (b).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
341,
999-0)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Todorovic,
and
B.R.Glick
(2008).
The interconversion of ACC deaminase and D: -cysteine desulfhydrase by directed mutagenesis.
|
| |
Planta,
229,
193-205.
|
 |
|
|
|
|
 |
Y.Tanaka,
K.Morikawa,
Y.Ohki,
M.Yao,
K.Tsumoto,
N.Watanabe,
T.Ohta,
and
I.Tanaka
(2007).
Structural and mutational analyses of Drp35 from Staphylococcus aureus: a possible mechanism for its lactonase activity.
|
| |
J Biol Chem,
282,
5770-5780.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Tanaka,
T.Sasaki,
I.Kumagai,
Y.Yasutake,
M.Yao,
I.Tanaka,
and
K.Tsumoto
(2007).
Molecular properties of two proteins homologous to PduO-type ATP:cob(I)alamin adenosyltransferase from Sulfolobus tokodaii.
|
| |
Proteins,
68,
446-457.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.R.Glick
(2005).
Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase.
|
| |
FEMS Microbiol Lett,
251,
1-7.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

