 |
PDBsum entry 1it7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine
|
|
Structure:
|
 |
Archaeosine tRNA-guanine transglycosylase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Pyrococcus horikoshii. Organism_taxid: 53953. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.229
|
R-free:
|
0.271
|
|
|
Authors:
|
 |
R.Ishitani,O.Nureki,S.Fukai,T.Kijimoto,N.Nameki,M.Watanabe,H.Kondo, M.Sekine,N.Okada,S.Nishimura,S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
R.Ishitani
et al.
(2002).
Crystal structure of archaeosine tRNA-guanine transglycosylase.
J Mol Biol,
318,
665-677.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Jan-02
|
Release date:
|
22-May-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O58843
(ATGT_PYRHO) -
tRNA-guanine(15) transglycosylase from Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
582 a.a.
577 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.48
- tRNA-guanine(15) transglycosylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
guanosine15 in tRNA + 7-cyano-7-deazaguanine = 7-cyano-7- carbaguanosine15 in tRNA + guanine
|
 |
 |
 |
 |
 |
guanosine(15) in tRNA
|
+
|
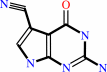 7-cyano-7-deazaguanine
7-cyano-7-deazaguanine
|
=
|
7-cyano-7- carbaguanosine(15) in tRNA
|
+
|
guanine
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
318:665-677
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of archaeosine tRNA-guanine transglycosylase.
|
|
R.Ishitani,
O.Nureki,
S.Fukai,
T.Kijimoto,
N.Nameki,
M.Watanabe,
H.Kondo,
M.Sekine,
N.Okada,
S.Nishimura,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Archaeosine tRNA-guanine transglycosylase (ArcTGT) catalyzes the exchange of
guanine at position 15 in the D-loop of archaeal tRNAs with a free
7-cyano-7-deazaguanine (preQ(0)) base, as the first step in the biosynthesis of
an archaea-specific modified base, archaeosine (7-formamidino-7-deazaguanosine).
We determined the crystal structures of ArcTGT from Pyrococcus horikoshii at 2.2
A resolution and its complexes with guanine and preQ(0), at 2.3 and 2.5 A
resolutions, respectively. The N-terminal catalytic domain folds into an
(alpha/beta)(8) barrel with a characteristic zinc-binding site, showing
structural similarity with that of the bacterial queuosine TGT (QueTGT), which
is involved in queuosine
(7-[[(4,5-cis-dihydroxy-2-cyclopenten-1-yl)-amino]methyl]-7-deazaguanosine)
biosynthesis and targets the tRNA anticodon. ArcTGT forms a dimer, involving the
zinc-binding site and the ArcTGT-specific C-terminal domain. The C-terminal
domains have novel folds, including an OB fold-like "PUA domain",
whose sequence is widely conserved in eukaryotic and archaeal RNA modification
enzymes. Therefore, the C-terminal domains may be involved in tRNA recognition.
In the free-form structure of ArcTGT, an alpha-helix located at the rim of the
(alpha/beta)(8) barrel structure is completely disordered, while it is ordered
in the guanine-bound and preQ(0)-bound forms. Structural comparison of the
ArcTGT.preQ(0), ArcTGT.guanine, and QueTGT.preQ(1) complexes provides novel
insights into the substrate recognition mechanisms of ArcTGT.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Domain architecture of the ArcTGT subunit. (a)
Ribbon diagram of the ArcTGT subunit. The catalytic domain,
domains C1, C2, and C3 are colored yellow, emerald green, sky
blue, and dark blue, respectively. Zinc and magnesium ions are
shown as metallic balls. (b) Topology diagram of the ArcTGT
structure. a-Helices are represented with circles or tubes, and
b-strands are shown with rectangles or arrows.
|
 |
Figure 7.
Figure 7. The catalytic domain of the preQ[0]-bound form of
(a) ArcTGT and (b) the base-free form of QueTGT. The viewpoints
of (a) and (b) are adjusted to that of Figure 6. The color of
helix a5 corresponds to the atomic B-factors, as indicated
below. The arrows indicate the major structural differences
between the ArcTGT and QueTGT, which are discussed in the text.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
318,
665-677)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.C.Chen,
A.F.Brooks,
D.M.Goodenough-Lashua,
J.D.Kittendorf,
H.D.Showalter,
and
G.A.Garcia
(2011).
Evolution of eukaryal tRNA-guanine transglycosylase: insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases.
|
| |
Nucleic Acids Res,
39,
2834-2844.
|
 |
|
|
|
|
 |
J.S.Luz,
C.R.Ramos,
M.C.Santos,
P.P.Coltri,
F.L.Palhano,
D.Foguel,
N.I.Zanchin,
and
C.C.Oliveira
(2010).
Identification of archaeal proteins that affect the exosome function in vitro.
|
| |
BMC Biochem,
11,
22.
|
 |
|
|
|
|
 |
M.Vinayak,
and
C.Pathak
(2010).
Queuosine modification of tRNA: its divergent role in cellular machinery.
|
| |
Biosci Rep,
30,
135-148.
|
 |
|
|
|
|
 |
X.Ming,
and
F.Seela
(2010).
Efficient synthesis of the tRNA nucleoside preQ0, 7-cyano-7-deazaguanosine, via microwave-assisted iodo→carbonitrile exchange.
|
| |
Chem Biodivers,
7,
2616-2621.
|
 |
|
|
|
|
 |
B.E.Yacoubi,
G.Phillips,
I.K.Blaby,
C.E.Haas,
Y.Cruz,
J.Greenberg,
and
V.de Crécy-Lagard
(2009).
A Gateway platform for functional genomics in Haloferax volcanii: deletion of three tRNA modification genes.
|
| |
Archaea,
2,
211-219.
|
 |
|
|
|
|
 |
C.Boland,
P.Hayes,
I.Santa-Maria,
S.Nishimura,
and
V.P.Kelly
(2009).
Queuosine Formation in Eukaryotic tRNA Occurs via a Mitochondria-localized Heteromeric Transglycosylase.
|
| |
J Biol Chem,
284,
18218-18227.
|
 |
|
|
|
|
 |
S.Muller,
A.Urban,
A.Hecker,
F.Leclerc,
C.Branlant,
and
Y.Motorin
(2009).
Deficiency of the tRNATyr:Psi 35-synthase aPus7 in Archaea of the Sulfolobales order might be rescued by the H/ACA sRNA-guided machinery.
|
| |
Nucleic Acids Res,
37,
1308-1322.
|
 |
|
|
|
|
 |
H.Takagi
(2008).
Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications.
|
| |
Appl Microbiol Biotechnol,
81,
211-223.
|
 |
|
|
|
|
 |
K.Miyazono,
Y.Nishimura,
Y.Sawano,
T.Makino,
and
M.Tanokura
(2008).
Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
|
| |
Proteins,
73,
1068-1071.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Gao,
and
R.S.Gupta
(2007).
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.
|
| |
BMC Genomics,
8,
86.
|
 |
|
|
|
|
 |
I.Pérez-Arellano,
J.Gallego,
and
J.Cervera
(2007).
The PUA domain - a structural and functional overview.
|
| |
FEBS J,
274,
4972-4984.
|
 |
|
|
|
|
 |
J.Payandeh,
and
E.F.Pai
(2007).
Enzyme-driven speciation: crystallizing Archaea via lipid capture.
|
| |
J Mol Evol,
64,
364-374.
|
 |
|
|
|
|
 |
N.Tidten,
B.Stengl,
A.Heine,
G.A.Garcia,
G.Klebe,
and
K.Reuter
(2007).
Glutamate versus glutamine exchange swaps substrate selectivity in tRNA-guanine transglycosylase: insight into the regulation of substrate selectivity by kinetic and crystallographic studies.
|
| |
J Mol Biol,
374,
764-776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.de Crécy-Lagard
(2007).
Identification of genes encoding tRNA modification enzymes by comparative genomics.
|
| |
Methods Enzymol,
425,
153-183.
|
 |
|
|
|
|
 |
J.Sabina,
and
D.Söll
(2006).
The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation.
|
| |
J Biol Chem,
281,
6993-7001.
|
 |
|
|
|
|
 |
L.M.Iyer,
A.M.Burroughs,
and
L.Aravind
(2006).
The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism.
|
| |
Bioinformatics,
22,
257-263.
|
 |
|
|
|
|
 |
X.Manival,
C.Charron,
J.B.Fourmann,
F.Godard,
B.Charpentier,
and
C.Branlant
(2006).
Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity.
|
| |
Nucleic Acids Res,
34,
826-839.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Stengl,
K.Reuter,
and
G.Klebe
(2005).
Mechanism and substrate specificity of tRNA-guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism.
|
| |
Chembiochem,
6,
1926-1939.
|
 |
|
|
|
|
 |
G.A.Garcia,
and
J.D.Kittendorf
(2005).
Transglycosylation: a mechanism for RNA modification (and editing?).
|
| |
Bioorg Chem,
33,
229-251.
|
 |
|
|
|
|
 |
K.A.Todorov,
X.J.Tan,
S.T.Nonekowski,
G.A.Garcia,
and
H.A.Carlson
(2005).
The role of aspartic acid 143 in E. coli tRNA-guanine transglycosylase: insights from mutagenesis studies and computational modeling.
|
| |
Biophys J,
89,
1965-1977.
|
 |
|
|
|
|
 |
S.Yang,
R.F.Doolittle,
and
P.E.Bourne
(2005).
Phylogeny determined by protein domain content.
|
| |
Proc Natl Acad Sci U S A,
102,
373-378.
|
 |
|
|
|
|
 |
W.Sun,
X.Xu,
M.Pavlova,
A.M.Edwards,
A.Joachimiak,
A.Savchenko,
and
D.Christendat
(2005).
The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
|
| |
Protein Sci,
14,
3121-3128.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Pérez-Arellano,
F.Gil-Ortiz,
J.Cervera,
and
V.Rubio
(2004).
Glutamate-5-kinase from Escherichia coli: gene cloning, overexpression, purification and crystallization of the recombinant enzyme and preliminary X-ray studies.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2091-2094.
|
 |
|
|
|
|
 |
A.R.Ferré-D'Amaré
(2003).
RNA-modifying enzymes.
|
| |
Curr Opin Struct Biol,
13,
49-55.
|
 |
|
|
|
|
 |
H.Hori,
S.Kubota,
K.Watanabe,
J.M.Kim,
T.Ogasawara,
T.Sawasaki,
and
Y.Endo
(2003).
Aquifex aeolicus tRNA (Gm18) methyltransferase has unique substrate specificity. TRNA recognition mechanism of the enzyme.
|
| |
J Biol Chem,
278,
25081-25090.
|
 |
|
|
|
|
 |
R.Brenk,
M.T.Stubbs,
A.Heine,
K.Reuter,
and
G.Klebe
(2003).
Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase and their implications for substrate selectivity, reaction mechanism and structure-based drug design.
|
| |
Chembiochem,
4,
1066-1077.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Ishitani,
O.Nureki,
N.Nameki,
N.Okada,
S.Nishimura,
and
S.Yokoyama
(2003).
Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme.
|
| |
Cell,
113,
383-394.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Xie,
X.Liu,
and
R.H.Huang
(2003).
Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate.
|
| |
Nat Struct Biol,
10,
781-788.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

