 |
PDBsum entry 1q2s

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/RNA
|
PDB id
|
|

|

|
1q2s
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.29
- tRNA-guanosine(34) preQ1 transglycosylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
7-aminomethyl-7-carbaguanine + guanosine34 in tRNA = 7-aminomethyl-7- carbaguanosine34 in tRNA + guanine
|
 |
 |
 |
 |
 |
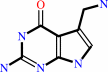 7-aminomethyl-7-carbaguanine
7-aminomethyl-7-carbaguanine
|
+
|
guanosine(34) in tRNA
|
=
|
7-aminomethyl-7- carbaguanosine(34) in tRNA
|
+
|
guanine
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
10:781-788
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate.
|
|
W.Xie,
X.Liu,
R.H.Huang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prokaryotic tRNA guanine transglycosylase (TGT) catalyzes replacement of guanine
(G) by 7-aminomethyl-7-deazaguanine (PreQ1) at the wobble position of four
specific tRNAs. Addition of 9-deazaguanine (9dzG) to a reaction mixture of
Zymomonas mobilis TGT and an RNA substrate allowed us to trap, purify and
crystallize a chemically competent covalent intermediate of the TGT-catalyzed
reaction. The crystal structure of the TGT-RNA-9dzG ternary complex at a
resolution of 2.9 A reveals, unexpectedly, that RNA is tethered to TGT through
the side chain of Asp280. Thus, Asp280, instead of the previously proposed
Asp102, acts as the nucleophile for the reaction. The RNA substrate adopts an
unusual conformation, with four out of seven nucleotides in the loop region
flipped out. Interactions between TGT and RNA revealed by the structure provide
the molecular basis of the RNA substrate requirements by TGT. Furthermore,
reaction of PreQ1 with the crystallized covalent intermediate provides insight
into the necessary structural changes required for the TGT-catalyzed reaction to
occur.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Overall structure of the trapped covalent
intermediate. (a) Ribbon representation of the TGT -RNA -9dzG
ternary complex. TGT is colored according to the assignment of
the secondary structures in Figure 2. The RNA is dark green.
9dzG is shown in ball-and-stick model in red, and a zinc ion is
represented by a ball in magenta. (b) Summary of TGT -RNA, TGT
-9dzG and RNA -RNA interactions. TGT, RNA and 9dzG are colored
as in a except the phosphate groups of RNA are in magenta. The
conserved RNA nucleotides 33 -35, 9dzG and Asp280 are
highlighted with color-filled boxes. Arrows in blue represent
hydrogen bonds between RNA and TGT, RNA and water, or 9dzG and
TGT. Arrows in red show stacking of amino acid side chains from
TGT on bases of RNA or on 9dzG. Two arrows in green represent
intramolecular interactions in RNA. 'W' in a circle represents
well-positioned water molecules in the structure.
|
 |
Figure 5.
Figure 5. The active site and molecular basis of substrate
specificity. (a) Stereo view of TGT active site. The main
chains of TGT are grayish blue, the side chains of TGT are
orange and the RNA and 9dzG are green. The hetero-atoms are
colored individually, with nitrogen in blue, oxygen in red and
phosphate in magenta. The C9 of 9dzG is highlighted in black. A
water molecule is shown as a small ball in red and labeled 'W'.
Hydrogen bonds are indicated with dashed lines. The side chains
of Asp106 and Cys158, which block 9dzG from the top, are omitted
for clarity. (b,c) Recognition of U33 (b) and U35 (c) in RNA
substrate by TGT. The color schemes are the same as in a. The
structure of the phosphate group of A38 in c is omitted but
labeled for clarity.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2003,
10,
781-788)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Guelorget,
and
B.Golinelli-Pimpaneau
(2011).
Mechanism-based strategies for trapping and crystallizing complexes of RNA-modifying enzymes.
|
| |
Structure,
19,
282-291.
|
 |
|
|
|
|
 |
S.Hebecker,
W.Arendt,
I.U.Heinemann,
J.H.Tiefenau,
M.Nimtz,
M.Rohde,
D.Söll,
and
J.Moser
(2011).
Alanyl-phosphatidylglycerol synthase: mechanism of substrate recognition during tRNA-dependent lipid modification in Pseudomonas aeruginosa.
|
| |
Mol Microbiol,
80,
935-950.
|
 |
|
|
|
|
 |
Y.C.Chen,
A.F.Brooks,
D.M.Goodenough-Lashua,
J.D.Kittendorf,
H.D.Showalter,
and
G.A.Garcia
(2011).
Evolution of eukaryal tRNA-guanine transglycosylase: insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases.
|
| |
Nucleic Acids Res,
39,
2834-2844.
|
 |
|
|
|
|
 |
M.Vinayak,
and
C.Pathak
(2010).
Queuosine modification of tRNA: its divergent role in cellular machinery.
|
| |
Biosci Rep,
30,
135-148.
|
 |
|
|
|
|
 |
Y.C.Chen,
V.P.Kelly,
S.V.Stachura,
and
G.A.Garcia
(2010).
Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure.
|
| |
RNA,
16,
958-968.
|
 |
|
|
|
|
 |
C.Boland,
P.Hayes,
I.Santa-Maria,
S.Nishimura,
and
V.P.Kelly
(2009).
Queuosine Formation in Eukaryotic tRNA Occurs via a Mitochondria-localized Heteromeric Transglycosylase.
|
| |
J Biol Chem,
284,
18218-18227.
|
 |
|
|
|
|
 |
G.A.Garcia,
S.M.Chervin,
and
J.D.Kittendorf
(2009).
Identification of the rate-determining step of tRNA-guanine transglycosylase from Escherichia coli.
|
| |
Biochemistry,
48,
11243-11251.
|
 |
|
|
|
|
 |
K.Nakanishi,
L.Bonnefond,
S.Kimura,
T.Suzuki,
R.Ishitani,
and
O.Nureki
(2009).
Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
|
| |
Nature,
461,
1144-1148.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Chimnaronk,
F.Forouhar,
J.Sakai,
M.Yao,
C.M.Tron,
M.Atta,
M.Fontecave,
J.F.Hunt,
and
I.Tanaka
(2009).
Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon.
|
| |
Biochemistry,
48,
5057-5065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Goto-Ito,
T.Ito,
M.Kuratani,
Y.Bessho,
and
S.Yokoyama
(2009).
Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation.
|
| |
Nat Struct Mol Biol,
16,
1109-1115.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Ritschel,
P.C.Kohler,
G.Neudert,
A.Heine,
F.Diederich,
and
G.Klebe
(2009).
How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase.
|
| |
ChemMedChem,
4,
2012-2023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Ritschel,
S.Hoertner,
A.Heine,
F.Diederich,
and
G.Klebe
(2009).
Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT.
|
| |
Chembiochem,
10,
716-727.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Cicmil,
and
L.Shi
(2008).
Crystallization and preliminary X-ray characterization of queD from Bacillus subtilis, an enzyme involved in queuosine biosynthesis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
119-122.
|
 |
|
|
|
|
 |
N.Cicmil,
and
R.H.Huang
(2008).
Crystal structure of QueC from Bacillus subtilis: an enzyme involved in preQ1 biosynthesis.
|
| |
Proteins,
72,
1084-1088.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Li
(2007).
Complexes of tRNA and maturation enzymes: shaping up for translation.
|
| |
Curr Opin Struct Biol,
17,
293-301.
|
 |
|
|
|
|
 |
J.K.Hurt,
S.Olgen,
and
G.A.Garcia
(2007).
Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro.
|
| |
Nucleic Acids Res,
35,
4905-4913.
|
 |
|
|
|
|
 |
N.Tidten,
B.Stengl,
A.Heine,
G.A.Garcia,
G.Klebe,
and
K.Reuter
(2007).
Glutamate versus glutamine exchange swaps substrate selectivity in tRNA-guanine transglycosylase: insight into the regulation of substrate selectivity by kinetic and crystallographic studies.
|
| |
J Mol Biol,
374,
764-776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.M.Chervin,
J.D.Kittendorf,
and
G.A.Garcia
(2007).
Probing the intermediacy of covalent RNA enzyme complexes in RNA modification enzymes.
|
| |
Methods Enzymol,
425,
121-137.
|
 |
|
|
|
|
 |
W.Xie,
C.Zhou,
and
R.H.Huang
(2007).
Structure of tRNA dimethylallyltransferase: RNA modification through a channel.
|
| |
J Mol Biol,
367,
872-881.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.C.Losey,
A.J.Ruthenburg,
and
G.L.Verdine
(2006).
Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA.
|
| |
Nat Struct Mol Biol,
13,
153-159.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Sabina,
and
D.Söll
(2006).
The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation.
|
| |
J Biol Chem,
281,
6993-7001.
|
 |
|
|
|
|
 |
B.Stengl,
K.Reuter,
and
G.Klebe
(2005).
Mechanism and substrate specificity of tRNA-guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism.
|
| |
Chembiochem,
6,
1926-1939.
|
 |
|
|
|
|
 |
G.A.Garcia,
and
J.D.Kittendorf
(2005).
Transglycosylation: a mechanism for RNA modification (and editing?).
|
| |
Bioorg Chem,
33,
229-251.
|
 |
|
|
|
|
 |
K.A.Todorov,
X.J.Tan,
S.T.Nonekowski,
G.A.Garcia,
and
H.A.Carlson
(2005).
The role of aspartic acid 143 in E. coli tRNA-guanine transglycosylase: insights from mutagenesis studies and computational modeling.
|
| |
Biophys J,
89,
1965-1977.
|
 |
|
|
|
|
 |
H.Okamoto,
K.Watanabe,
Y.Ikeuchi,
T.Suzuki,
Y.Endo,
and
H.Hori
(2004).
Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus.
|
| |
J Biol Chem,
279,
49151-49159.
|
 |
|
|
|
|
 |
K.Phannachet,
and
R.H.Huang
(2004).
Conformational change of pseudouridine 55 synthase upon its association with RNA substrate.
|
| |
Nucleic Acids Res,
32,
1422-1429.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Del Campo,
J.Ofengand,
and
A.Malhotra
(2004).
Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli.
|
| |
RNA,
10,
231-239.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Kaya,
M.Del Campo,
J.Ofengand,
and
A.Malhotra
(2004).
Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold.
|
| |
J Biol Chem,
279,
18107-18110.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

