 |
PDBsum entry 1hkv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Mycobacterium diaminopimelate dicarboxylase (lysa)
|
|
Structure:
|
 |
Diaminopimelate decarboxylase. Chain: a, b. Synonym: dap decarboxylase, meso-diaminopimelate decarboxylase, lysa. Engineered: yes. Other_details: complexed with cofactor plp and product lysine
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 83332. Strain: h37rv. Expressed in: escherichia coli. Expression_system_taxid: 562. Other_details: c-term 6-his tag
|
|
Biol. unit:
|
 |
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.227
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
K.Gokulan,B.Rupp,M.S.Pavelka Jr,W.R.Jacobs Jr,J.C.Sacchettini,Tb Structural Genomics Consortium (Tbsgc)
|
Key ref:

|
 |
K.Gokulan
et al.
(2003).
Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis.
J Biol Chem,
278,
18588-18596.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Mar-03
|
Release date:
|
20-Mar-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WIU7
(DCDA_MYCTU) -
Diaminopimelate decarboxylase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
447 a.a.
446 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.20
- diaminopimelate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine biosynthesis (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
meso-2,6-diaminopimelate + H+ = L-lysine + CO2
|
 |
 |
 |
 |
 |
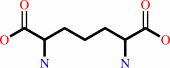 meso-2,6-diaminopimelate
meso-2,6-diaminopimelate
|
+
|
H(+)
|
=
|
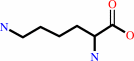 L-lysine
L-lysine
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:18588-18596
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis.
|
|
K.Gokulan,
B.Rupp,
M.S.Pavelka,
W.R.Jacobs,
J.C.Sacchettini.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Mycobacterium tuberculosis lysA gene encodes the enzyme meso-diaminopimelate
decarboxylase (DAPDC), a pyridoxal-5'-phosphate (PLP)-dependent enzyme. The
enzyme catalyzes the final step in the lysine biosynthetic pathway converting
meso-diaminopimelic acid (DAP) to l-lysine. The lysA gene of M. tuberculosis
H37Rv has been established as essential for bacterial survival in
immunocompromised mice, demonstrating that de novo biosynthesis of lysine is
essential for in vivo viability. Drugs targeted against DAPDC could be efficient
anti-tuberculosis drugs, and the three-dimensional structure of DAPDC from M.
tuberculosis complexed with reaction product lysine and the ternary complex with
PLP and lysine in the active site has been determined. The first structure of a
DAPDC confirms its classification as a fold type III PLP-dependent enzyme. The
structure shows a stable 2-fold dimer in head-to-tail arrangement of a
triose-phosphate isomerase (TIM) barrel-like alpha/beta domain and a C-terminal
beta sheet domain, similar to the ornithine decarboxylase (ODC) fold family. PLP
is covalently bound via an internal aldimine, and residues from both domains and
both subunits contribute to the binding pocket. Comparison of the structure with
eukaryotic ODCs, in particular with a di-fluoromethyl ornithine (DMFO)-bound ODC
from Trypanosoma bruceii, indicates that corresponding DAP-analogues might be
potential inhibitors for mycobacterial DAPDCs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Multiple sequence alignment of PLP-dependent
enzymes. Top line indicates regions of partially conserved or
important binding motives or residues. Alignment carried out
with ClustalW 1.8.2 (40). Color key: green, polar residues; red,
hydrophobic residues; blue, negatively charged; and magenta,
positively charged.
|
 |
Figure 6.
Fig. 6. Schematic representation of ligand binding
interactions in active site pocket of DAPDC. Residues of both
homodimer subunits contribute to PLP and to lysine binding. This
figure was created by LIGPLOT (43).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
18588-18596)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Muramatsu,
Y.Suzuki,
T.Imai,
S.Ueshima,
J.Ozaki,
Y.Matsui,
S.Kato,
K.Ohnishi,
N.Kimoto,
H.Yamamoto,
and
S.Nagata
(2011).
Discovery and characterization of D: -phenylserine deaminase from Arthrobacter sp. TKS1.
|
| |
Appl Microbiol Biotechnol,
90,
159-172.
|
 |
|
|
|
|
 |
S.Weyand,
G.Kefala,
D.I.Svergun,
and
M.S.Weiss
(2009).
The three-dimensional structure of diaminopimelate decarboxylase from Mycobacterium tuberculosis reveals a tetrameric enzyme organisation.
|
| |
J Struct Funct Genomics,
10,
209-217.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Raman,
Y.Kalidas,
and
N.Chandra
(2008).
targetTB: A target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis.
|
| |
BMC Syst Biol,
2,
109.
|
 |
|
|
|
|
 |
C.A.Hutton,
M.A.Perugini,
and
J.A.Gerrard
(2007).
Inhibition of lysine biosynthesis: an evolving antibiotic strategy.
|
| |
Mol Biosyst,
3,
458-465.
|
 |
|
|
|
|
 |
R.Shah,
R.Akella,
E.J.Goldsmith,
and
M.A.Phillips
(2007).
X-ray structure of Paramecium bursaria Chlorella virus arginine decarboxylase: insight into the structural basis for substrate specificity.
|
| |
Biochemistry,
46,
2831-2841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Alexeev,
R.L.Baxter,
D.J.Campopiano,
O.Kerbarh,
L.Sawyer,
N.Tomczyk,
R.Watt,
and
S.P.Webster
(2006).
Suicide inhibition of alpha-oxamine synthases: structures of the covalent adducts of 8-amino-7-oxononanoate synthase with trifluoroalanine.
|
| |
Org Biomol Chem,
4,
1209-1212.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.A.Azevedo,
M.Lancien,
and
P.J.Lea
(2006).
The aspartic acid metabolic pathway, an exciting and essential pathway in plants.
|
| |
Amino Acids,
30,
143-162.
|
 |
|
|
|
|
 |
S.Hasan,
S.Daugelat,
P.S.Rao,
and
M.Schreiber
(2006).
Prioritizing genomic drug targets in pathogens: application to Mycobacterium tuberculosis.
|
| |
PLoS Comput Biol,
2,
e61.
|
 |
|
|
|
|
 |
S.Jantaro,
H.Kidron,
D.Chesnel,
A.Incharoensakdi,
P.Mulo,
T.Salminen,
and
P.Mäenpää
(2006).
Structural modeling and environmental regulation of arginine decarboxylase in Synechocystis sp. PCC 6803.
|
| |
Arch Microbiol,
184,
397-406.
|
 |
|
|
|
|
 |
V.L.Arcus,
J.S.Lott,
J.M.Johnston,
and
E.N.Baker
(2006).
The potential impact of structural genomics on tuberculosis drug discovery.
|
| |
Drug Discov Today,
11,
28-34.
|
 |
|
|
|
|
 |
G.Kefala,
L.J.Perry,
and
M.S.Weiss
(2005).
Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of LysA (Rv1293) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
782-784.
|
 |
|
|
|
|
 |
C.N.Patel,
R.S.Adcock,
K.G.Sell,
and
M.A.Oliveira
(2004).
Crystallization, X-ray diffraction and oligomeric characterization of arginine decarboxylase from Yersinia pestis, a key polyamine biosynthetic enzyme.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2396-2398.
|
 |
|
|
|
|
 |
P.R.Hall,
R.Zheng,
L.Antony,
M.Pusztai-Carey,
P.R.Carey,
and
V.C.Yee
(2004).
Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
|
| |
EMBO J,
23,
3621-3631.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
M.Bellinzoni,
and
G.Riccardi
(2003).
Techniques and applications: The heterologous expression of Mycobacterium tuberculosis genes is an uphill road.
|
| |
Trends Microbiol,
11,
351-358.
|
 |
|
|
|
|
 |
P.B.Balbo,
C.N.Patel,
K.G.Sell,
R.S.Adcock,
S.Neelakantan,
P.A.Crooks,
and
M.A.Oliveira
(2003).
Spectrophotometric and steady-state kinetic analysis of the biosynthetic arginine decarboxylase of Yersinia pestis utilizing arginine analogues as inhibitors and alternative substrates.
|
| |
Biochemistry,
42,
15189-15196.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

