 |
PDBsum entry 1g8h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|

|

|

|

|

|
 _CD
×11 _CD
×11
|
 |

|

|

|

|

|

|

|
 _CA
×6 _CA
×6
|
 |

|

|

|

|

|

|

|
 _NA
×12 _NA
×12
|
 |

|

|

|

|

|

|

|
 _MG
×2 _MG
×2
|
 |

|

|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.4
- sulfate adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
sulfate + ATP + H+ = adenosine 5'-phosphosulfate + diphosphate
|
 |
 |
 |
 |
 |
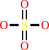 sulfate
sulfate
|
+
|
 ATP
ATP
|
+
|
H(+)
|
=
|
adenosine 5'-phosphosulfate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
diphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
20:316-329
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
|
|
T.C.Ullrich,
M.Blaesse,
R.Huber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
ATP sulfurylases (ATPSs) are ubiquitous enzymes that catalyse the primary step
of intracellular sulfate activation: the reaction of inorganic sulfate with ATP
to form adenosine-5'-phosphosulfate (APS) and pyrophosphate (PPi). With the
crystal structure of ATPS from the yeast Saccharomyces cerevisiae, we have
solved the first structure of a member of the ATP sulfurylase family. We have
analysed the crystal structure of the native enzyme at 1.95 Angstroms resolution
using multiple isomorphous replacement (MIR) and, subsequently, the ternary
enzyme product complex with APS and PPi bound to the active site. The enzyme
consists of six identical subunits arranged in two stacked rings in a D:3
symmetric assembly. Nucleotide binding causes significant conformational
changes, which lead to a rigid body structural displacement of domains III and
IV of the ATPS monomer. Despite having similar folds and active site design,
examination of the active site of ATPS and comparison with known structures of
related nucleotidylyl transferases reveal a novel ATP binding mode that is
peculiar to ATP sulfurylases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5 Active site residues of the apo-protomer liganded with
sulfate at 1.95 Å (A), the binary enzyme product complex with
APS at 2.6 Å (B), and the ternary product complex with an
additional pyrophosphate at 2.8 Å (C). All atoms are shown with
the final 2F[o] - F[c] electron density contoured at 1.0  . .
|
 |
Figure 6.
Figure 6 Schematic drawing of the coordination of APS in the
binding pocket, its hydrogen bonding and hydrophobic
interactions with solvent molecules and the residues of ATPS.
Bond distances are given in Å.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2001,
20,
316-329)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Bertonati,
M.Punta,
M.Fischer,
G.Yachdav,
F.Forouhar,
W.Zhou,
A.P.Kuzin,
J.Seetharaman,
M.Abashidze,
T.A.Ramelot,
M.A.Kennedy,
J.R.Cort,
A.Belachew,
J.F.Hunt,
L.Tong,
G.T.Montelione,
and
B.Rost
(2009).
Structural genomics reveals EVE as a new ASCH/PUA-related domain.
|
| |
Proteins,
75,
760-773.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Bradley,
J.S.Rest,
W.H.Li,
and
N.B.Schwartz
(2009).
Sulfate activation enzymes: phylogeny and association with pyrophosphatase.
|
| |
J Mol Evol,
68,
1.
|
 |
|
|
|
|
 |
S.C.Gay,
I.H.Segel,
and
A.J.Fisher
(2009).
Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: structural basis for the absence of ATP sulfurylase activity.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1021-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Y.Gavel,
A.V.Kladova,
S.A.Bursakov,
J.M.Dias,
S.Texeira,
V.L.Shnyrov,
J.J.Moura,
I.Moura,
M.J.Romão,
and
J.Trincão
(2008).
Purification, crystallization and preliminary X-ray diffraction analysis of adenosine triphosphate sulfurylase (ATPS) from the sulfate-reducing bacterium Desulfovibrio desulfuricans ATCC 27774.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
593-595.
|
 |
|
|
|
|
 |
T.Simonics,
and
A.Maráz
(2008).
Cloning of the ATP sulphurylase gene of Schizosaccharomyces pombe by functional complementation.
|
| |
Can J Microbiol,
54,
71-74.
|
 |
|
|
|
|
 |
J.D.Mougous,
D.H.Lee,
S.C.Hubbard,
M.W.Schelle,
D.J.Vocadlo,
J.M.Berger,
and
C.R.Bertozzi
(2006).
Molecular basis for G protein control of the prokaryotic ATP sulfurylase.
|
| |
Mol Cell,
21,
109-122.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.W.Schelle,
and
C.R.Bertozzi
(2006).
Sulfate metabolism in mycobacteria.
|
| |
Chembiochem,
7,
1516-1524.
|
 |
|
|
|
|
 |
T.Arakaki,
I.Le Trong,
E.Phizicky,
E.Quartley,
G.DeTitta,
J.Luft,
A.Lauricella,
L.Anderson,
O.Kalyuzhniy,
E.Worthey,
P.J.Myler,
D.Kim,
D.Baker,
W.G.Hol,
and
E.A.Merritt
(2006).
Structure of Lmaj006129AAA, a hypothetical protein from Leishmania major.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
175-179.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.D.Spiegelberg,
J.Dela Cruz,
T.H.Law,
and
J.D.York
(2005).
Alteration of lithium pharmacology through manipulation of phosphoadenosine phosphate metabolism.
|
| |
J Biol Chem,
280,
5400-5405.
|
 |
|
|
|
|
 |
D.Mendoza-Cózatl,
H.Loza-Tavera,
A.Hernández-Navarro,
and
R.Moreno-Sánchez
(2005).
Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants.
|
| |
FEMS Microbiol Rev,
29,
653-671.
|
 |
|
|
|
|
 |
W.Sun,
X.Xu,
M.Pavlova,
A.M.Edwards,
A.Joachimiak,
A.Savchenko,
and
D.Christendat
(2005).
The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
|
| |
Protein Sci,
14,
3121-3128.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Hanna,
K.F.Ng,
I.J.MacRae,
C.J.Bley,
A.J.Fisher,
and
I.H.Segel
(2004).
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory domain.
|
| |
J Biol Chem,
279,
4415-4424.
|
 |
|
|
|
|
 |
S.Harjes,
A.Scheidig,
and
P.Bayer
(2004).
Expression, purification and crystallization of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
350-352.
|
 |
|
|
|
|
 |
V.Saridakis,
and
E.F.Pai
(2003).
Mutational, structural, and kinetic studies of the ATP-binding site of Methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase.
|
| |
J Biol Chem,
278,
34356-34363.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Taguchi,
J.Hoseki,
Y.Kakuta,
and
K.Fukuyama
(2003).
Overproduction, crystallization and preliminary X-ray diffraction analysis of probable ATP sulfurylase from Thermus thermophilus HB8.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1645-1647.
|
 |
|
|
|
|
 |
I.J.MacRae,
I.H.Segel,
and
A.J.Fisher
(2002).
Allosteric inhibition via R-state destabilization in ATP sulfurylase from Penicillium chrysogenum.
|
| |
Nat Struct Biol,
9,
945-949.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Garavaglia,
I.D'Angelo,
M.Emanuelli,
F.Carnevali,
F.Pierella,
G.Magni,
and
M.Rizzi
(2002).
Structure of human NMN adenylyltransferase. A key nuclear enzyme for NAD homeostasis.
|
| |
J Biol Chem,
277,
8524-8530.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.J.MacRae,
I.H.Segel,
and
A.J.Fisher
(2001).
Crystal structure of ATP sulfurylase from Penicillium chrysogenum: insights into the allosteric regulation of sulfate assimilation.
|
| |
Biochemistry,
40,
6795-6804.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

