
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 155 a.a.
155 a.a.
|
 |

|

|

|

|

|

|

|

 797 a.a.
797 a.a.
|
 |

|

|

|

|

|

|

|

 287 a.a.
287 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Carbon monoxide dehydrogenase from hydrogenophaga pseudoflava
|
|
Structure:
|
 |
Cuts, iron-sulfur protein of carbon monoxide dehydrogenase. Chain: a, d. Cutl, molybdoprotein of carbon monoxide dehydrogenase. Chain: b, e. Cutm, flavoprotein of carbon monoxide dehydrogenase. Chain: c, f
|
|
Source:
|
 |
Hydrogenophaga pseudoflava. Organism_taxid: 47421. Organism_taxid: 47421
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.209
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
P.Haenzelmann,H.Dobbek,L.Gremer,R.Huber,O.Meyer
|
Key ref:

|
 |
P.Hänzelmann
et al.
(2000).
The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J Mol Biol,
301,
1221-1235.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Jul-00
|
Release date:
|
15-Sep-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P19915
(DCMS_HYDPS) -
Carbon monoxide dehydrogenase small chain from Hydrogenophaga pseudoflava
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
163 a.a.
155 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.2.5.3
- aerobic carbon monoxide dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
CO + a quinone + H2O = a quinol + CO2
|
 |
 |
 |
 |
 |
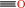 CO
CO
|
+
|
 quinone
quinone
|
+
|
H2O
|
=
|
quinol
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation; FAD; Iron-sulfur; Molybdenum atom; Molybdopterin
|
 |
 |
 |
 |
 |
Cu cation
|
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
 Iron-sulfur
Iron-sulfur
|
Molybdenum atom
|
Molybdopterin
Bound ligand (Het Group name =
PCD)
matches with 50.00% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
301:1221-1235
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
|
|
P.Hänzelmann,
H.Dobbek,
L.Gremer,
R.Huber,
O.Meyer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures of carbon monoxide dehydrogenase (CODH), a
seleno-molybdo-iron-sulfur flavoprotein from the aerobic carbon monoxide
utilizing carboxidotrophic eubacterium Hydrogenophaga pseudoflava, have been
determined from the enzyme synthesized at high (Mo(plus) CODH) and low
intracellular molybdenum content (Mo(minus) CODH) at 2.25 A and 2.35 A
resolution, respectively. The structures were solved by Patterson search methods
utilizing the enzyme from Oligotropha carboxidovorans as the initial model. The
CODHs from both sources are structurally very much conserved and show the same
overall fold, architecture and arrangements of the molybdopterin-cytosine
dinucleotide-type of molybdenum cofactor, the type I and type II [2Fe-2S]
clusters and the flavin-adenine dinucleotide. Unlike the CODH from O.
carboxidovorans, the enzyme from H. pseudoflava reveals a unique
post-translationally modified C(gamma)-hydroxy-Arg384 residue which precedes the
catalytically essential S-selanyl-Cys385 in the active-site loop. In addition,
the Trp193 which shields the isoalloxazine ring of the flavin-adenine
dinucleotide in the M subunit of the H. pseudoflava CODH is a Tyr193 in the O.
carboxidovorans CODH. The hydrogen bonding interaction pattern of the molybdenum
cofactor involves 27 hydrogen bonds with the surrounding protein. Of these,
eight are with the cytosine moiety, eight with the pyrophosphate, six with the
pyranopterin, and five with the ligands of the Mo ion. The structure of the
catalytically inactive Mo(minus) CODH indicates that an intracellular
Mo-deficiency affects exclusively the active site of the enzyme as an incomplete
non-functional molybdenum cofactor was synthesized. The 5'-CDP residue was
present in Mo(minus) CODH, whereas the Mo-pyranopterin moiety was absent. In
Mo(plus) CODH the selenium faces the Mo ion and flips away from the Mo site in
Mo(minus) CODH. The different side-chain conformations of the active-site
residues S-selanyl-Cys385 and Glu757 in Mo(plus) and Mo(minus) CODH indicate a
side-chain flexibility and a function of the Mo ion in the proper orientation of
both residues.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. The fold of the L-subunit of the Moplus CODH
species. (a) The C-terminal domain (residues 440 to 803). (b)
The N-terminal domain (residues 7 to 435). Helices are shown in
red, b-sheets in yellow and loops in green. Colour coding of the
Moco atoms is as follows: C, green; N, blue; O, red; P, pink.
The structure representations were created as in Figure 1.
|
 |
Figure 3.
Figure 3. Representation of the active sites of the Moplus
or the Mominus CODH species. (a) Moplus CODH. The Mo ion is
complexed by the enedithiolate group of MCD. It has one hydroxy
and two oxo ligands in the first coordination sphere. In the
second coordination sphere are the residues Gln237,
C^g-hydroxy-Arg384, S-selanyl-Cys385, Gly563 (not shown for
reasons of clarity) and Glu757. Hydrogen bonds are shown as
broken lines. Colour coding of the Moco atoms is as follows: C,
green; Mo, grey; N, blue; O, red; P, pink; S, yellow; Se,
orange. (b) Superposition of the active sites of the Moplus and
the Mominus CODH species. The active sites of the Moplus or
Mominus CODH species are drawn in green and red, respectively.
The structure representations were created as in Figure 1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
301,
1221-1235)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Neumann,
and
S.Leimkühler
(2011).
The role of system-specific molecular chaperones in the maturation of molybdoenzymes in bacteria.
|
| |
Biochem Res Int,
2011,
850924.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
N.Wagener,
A.J.Pierik,
A.Ibdah,
R.Hille,
and
H.Dobbek
(2009).
The Mo-Se active site of nicotinate dehydrogenase.
|
| |
Proc Natl Acad Sci U S A,
106,
11055-11060.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.D.Brondino,
M.J.Romão,
I.Moura,
and
J.J.Moura
(2006).
Molybdenum and tungsten enzymes: the xanthine oxidase family.
|
| |
Curr Opin Chem Biol,
10,
109-114.
|
 |
|
|
|
|
 |
L.Fieseler,
A.Quaiser,
C.Schleper,
and
U.Hentschel
(2006).
Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics.
|
| |
Environ Microbiol,
8,
612-624.
|
 |
|
|
|
|
 |
P.E.Siegbahn,
and
A.F.Shestakov
(2005).
Quantum chemical modeling of CO oxidation by the active site of molybdenum CO dehydrogenase.
|
| |
J Comput Chem,
26,
888-898.
|
 |
|
|
|
|
 |
V.Purvanov,
and
S.Fetzner
(2005).
Replacement of active-site residues of quinoline 2-oxidoreductase involved in substrate recognition and specificity.
|
| |
Curr Microbiol,
50,
217-222.
|
 |
|
|
|
|
 |
G.P.Roberts,
H.Youn,
and
R.L.Kerby
(2004).
CO-sensing mechanisms.
|
| |
Microbiol Mol Biol Rev,
68,
453-473.
|
 |
|
|
|
|
 |
I.Bonin,
B.M.Martins,
V.Purvanov,
S.Fetzner,
R.Huber,
and
H.Dobbek
(2004).
Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase.
|
| |
Structure,
12,
1425-1435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Unciuleac,
E.Warkentin,
C.C.Page,
M.Boll,
and
U.Ermler
(2004).
Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow.
|
| |
Structure,
12,
2249-2256.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Uchida,
D.Kondo,
A.Yamashita,
Y.Nagaosa,
T.Sakurai,
Y.Fujii,
K.Fujishiro,
K.Aisaka,
and
T.Uwajima
(2003).
Purification and characterization of an aldehyde oxidase from Pseudomonas sp. KY 4690.
|
| |
FEMS Microbiol Lett,
229,
31-36.
|
 |
|
|
|
|
 |
H.Dobbek,
L.Gremer,
R.Kiefersauer,
R.Huber,
and
O.Meyer
(2002).
Catalysis at a dinuclear [CuSMo(==O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution.
|
| |
Proc Natl Acad Sci U S A,
99,
15971-15976.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Dobbek,
V.Svetlitchnyi,
L.Gremer,
R.Huber,
and
O.Meyer
(2001).
Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster.
|
| |
Science,
293,
1281-1285.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Svetlitchnyi,
C.Peschel,
G.Acker,
and
O.Meyer
(2001).
Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans.
|
| |
J Bacteriol,
183,
5134-5144.
|
 |
|
|
|
|
 |
O.Meyer,
L.Gremer,
R.Ferner,
M.Ferner,
H.Dobbek,
M.Gnida,
W.Meyer-Klaucke,
and
R.Huber
(2000).
The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase.
|
| |
Biol Chem,
381,
865-876.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

