 |
PDBsum entry 1b80

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1b80
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.14
- lignin peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol + H2O2 = 3,4-dimethoxybenzaldehyde + guaiacol + glycolaldehyde + H2O
|
|
2.
|
2 (3,4-dimethoxyphenyl)methanol + H2O2 = 2 (3,4- dimethoxyphenyl)methanol radical + 2 H2O
|
|
 |
 |
 |
 |
 |
1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol
|
+
|
 H2O2
H2O2
|
=
|
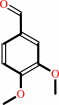 3,4-dimethoxybenzaldehyde
3,4-dimethoxybenzaldehyde
|
+
|
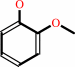 guaiacol
guaiacol
|
+
|
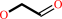 glycolaldehyde
glycolaldehyde
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
2
×
(3,4-dimethoxyphenyl)methanol
|
+
|
 H2O2
H2O2
|
=
|
2
×
(3,4- dimethoxyphenyl)methanol radical
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
305:851-861
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism.
|
|
W.Blodig,
A.T.Smith,
W.A.Doyle,
K.Piontek.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The heme enzyme lignin peroxidase (LiP) from the white rot fungus Phanerochaete
chrysosporium contains a solvent exposed redox active tryptophan residue
(Trp171) that carries a unique hydroxy group stereo-specifically attached to its
C(beta) atom. A Trp171Phe mutant has no activity at all towards the substrate
veratryl alcohol. The mechanism of veratryl alcohol oxidation involving
beta-hydroxy-Trp171 is largely unknown. Here, we present the first crystal
structures of LiP isozyme H8 at high resolution in its pristine non-hydroxylated
form, of the C(beta)-hydroxylated form, and of the Trp171Phe mutant using
recombinantly expressed and refolded protein produced from Escherichia coli. As
a consequence, all structures are unglycosylated. Structural changes in response
to the mutation are marginal and allow us to attribute the complete lack of
activity exclusively to the absence of the redox active indole side-chain. The
origin of the stereospecificity of the Trp171 hydroxylation can be explained on
structural grounds. A reaction mechanism involving Trp171 is proposed and the
possible function of the modification is discussed. Another important result
regarding the ongoing debate on the co-ordination state of the heme iron in the
resting state is that the iron is six co-ordinate in all cases the data being
collected at room temperature. The mean distance from the iron to the distal
water ligand is 2.18(+/-0.08) A. The radical scavenger orcinol was found to
decrease radiation damage to the crystals, during data collection at room
temperature.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Difference omit maps of (a) the
LiPH8-H[2]O[2]structure and (b) the pristine LiPH8 structure for
residue Trp171 at 1.73 and 1.8 Å resolution, respectively.
The maps in cyan are (2F[o] -F[c])exp(ia[c]) electron densities
contoured at 2 s where all atoms of Trp171 were omitted for
phase calculation. In red a (F[o] -F[c])exp(ia[c]) electron
density map is shown, contoured at 8 s where only the hydroxy
group was omitted. The picture was produced with O [Jones et al
1991].
|
 |
Figure 5.
Figure 5. Difference omit map ((F[o] -F[c])exp(ia[c])) at
1.85 Å resolution of the W171F mutant structure of
recombinant LiPH8 for residue Phe171 contoured at 7s. The water
molecule (Wat417) which hydrogen bonds to the hydroxy group at
the C^b atom of Trp171 in the LiP-H[2]O[2] structure is shown as
a red asterisk (see also Figure 4). The picture was produced
with O [Jones et al 1991].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
305,
851-861)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Bernini,
R.Pogni,
F.J.Ruiz-Dueñas,
A.T.Martínez,
R.Basosi,
and
A.Sinicropi
(2011).
EPR parameters of amino acid radicals in P. eryngii versatile peroxidase and its W164Y variant computed at the QM/MM level.
|
| |
Phys Chem Chem Phys,
13,
5078-5098.
|
 |
|
|
|
|
 |
M.Hofrichter,
R.Ullrich,
M.J.Pecyna,
C.Liers,
and
T.Lundell
(2010).
New and classic families of secreted fungal heme peroxidases.
|
| |
Appl Microbiol Biotechnol,
87,
871-897.
|
 |
|
|
|
|
 |
A.T.Smith,
W.A.Doyle,
P.Dorlet,
and
A.Ivancich
(2009).
Spectroscopic evidence for an engineered, catalytically active Trp radical that creates the unique reactivity of lignin peroxidase.
|
| |
Proc Natl Acad Sci U S A,
106,
16084-16089.
|
 |
|
|
|
|
 |
D.W.Wong
(2009).
Structure and action mechanism of ligninolytic enzymes.
|
| |
Appl Biochem Biotechnol,
157,
174-209.
|
 |
|
|
|
|
 |
F.J.Ruiz-Dueñas,
M.Morales,
E.García,
Y.Miki,
M.J.Martínez,
and
A.T.Martínez
(2009).
Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases.
|
| |
J Exp Bot,
60,
441-452.
|
 |
|
|
|
|
 |
F.J.Ruiz-Dueñas,
R.Pogni,
M.Morales,
S.Giansanti,
M.J.Mate,
A.Romero,
M.J.Martínez,
R.Basosi,
and
A.T.Martínez
(2009).
Protein Radicals in Fungal Versatile Peroxidase: CATALYTIC TRYPTOPHAN RADICAL IN BOTH COMPOUND I AND COMPOUND II AND STUDIES ON W164Y, W164H, AND W164S VARIANTS.
|
| |
J Biol Chem,
284,
7986-7994.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Morgenstern,
S.Klopman,
and
D.S.Hibbett
(2008).
Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes.
|
| |
J Mol Evol,
66,
243-257.
|
 |
|
|
|
|
 |
K.S.Hildén,
M.R.Mäkelä,
T.K.Hakala,
A.Hatakka,
and
T.Lundell
(2006).
Expression on wood, molecular cloning and characterization of three lignin peroxidase (LiP) encoding genes of the white rot fungus Phlebia radiata.
|
| |
Curr Genet,
49,
97.
|
 |
|
|
|
|
 |
R.Pogni,
M.C.Baratto,
C.Teutloff,
S.Giansanti,
F.J.Ruiz-Dueñas,
T.Choinowski,
K.Piontek,
A.T.Martínez,
F.Lendzian,
and
R.Basosi
(2006).
A tryptophan neutral radical in the oxidized state of versatile peroxidase from Pleurotus eryngii: a combined multifrequency EPR and density functional theory study.
|
| |
J Biol Chem,
281,
9517-9526.
|
 |
|
|
|
|
 |
C.Jung,
F.Lendzian,
V.Schünemann,
M.Richter,
L.H.Böttger,
A.X.Trautwein,
J.Contzen,
M.Galander,
D.K.Ghosh,
and
A.L.Barra
(2005).
Multi-frequency EPR and Mössbauer spectroscopic studies on freeze-quenched reaction intermediates of nitric oxide synthase.
|
| |
Magn Reson Chem,
43,
S84-S95.
|
 |
|
|
|
|
 |
C.Jung,
V.Schünemann,
F.Lendzian,
A.X.Trautwein,
J.Contzen,
M.Galander,
L.H.Böttger,
M.Richter,
and
A.L.Barra
(2005).
Spectroscopic characterization of the iron-oxo intermediate in cytochrome P450.
|
| |
Biol Chem,
386,
1043-1053.
|
 |
|
|
|
|
 |
Y.Sargisova,
F.M.Pierfederici,
A.Scirè,
E.Bertoli,
F.Tanfani,
F.Febbraio,
R.Briante,
Y.Karapetyan,
and
S.Mardanyan
(2004).
Computational, spectroscopic, and resonant mirror biosensor analysis of the interaction of adrenodoxin with native and tryptophan-modified NADPH-adrenodoxin reductase.
|
| |
Proteins,
57,
302-310.
|
 |
|
|
|
|
 |
G.Ward,
Y.Hadar,
I.Bilkis,
and
C.G.Dosoretz
(2003).
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes.
|
| |
J Biol Chem,
278,
39726-39734.
|
 |
|
|
|
|
 |
M.Francesca Gerini,
D.Roccatano,
E.Baciocchi,
and
A.Di Nola
(2003).
Molecular dynamics simulations of lignin peroxidase in solution.
|
| |
Biophys J,
84,
3883-3893.
|
 |
|
|
|
|
 |
O.M.Lardinois,
and
P.R.Ortiz de Montellano
(2003).
Intra- and intermolecular transfers of protein radicals in the reactions of sperm whale myoglobin with hydrogen peroxide.
|
| |
J Biol Chem,
278,
36214-36226.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

