 |
PDBsum entry 1an9

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1an9
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.3
- D-amino-acid oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Cephalosporin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a D-alpha-amino acid + O2 + H2O = a 2-oxocarboxylate + H2O2 + NH4+
|
 |
 |
 |
 |
 |
D-alpha-amino acid
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
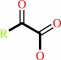 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
122:825-833
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
|
|
R.Miura,
C.Setoyama,
Y.Nishina,
K.Shiga,
H.Mizutani,
I.Miyahara,
K.Hirotsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
As an extension of our recent X-ray crystallographic determination of the
tertiary structure of D-amino acid oxidase (DAO) [Mizutani, H. et al. (1996) J.
Biochem. 120, 14-17], we solved the crystal structure of the complex of DAO with
a substrate analog, o-aminobenzoate (OAB). The alignment between flavin and OAB
in the crystal structure of the complex is consistent with charge-transfer
interaction through the overlap between the highest occupied molecular orbital
of OAB and the lowest unoccupied molecular orbital of flavin. Starting with the
atomic coordinates of this complex as the initial model, we carried out
molecular mechanics simulation for the DAO-D-leucine complex and thus obtained a
model for the enzyme-substrate complex. According to the enzyme-substrate
complex model, the alpha-proton is pointed toward N(5) of flavin while the
lone-pair of the substrate amino group can approach C(4a) of flavin within an
interacting distance. This model as well as DAO-OAB complex enables the
evaluation of the substrate-flavin interaction prior to electron transfer from
the substrate to flavin and provides two possible mechanisms for the
reductive-half reaction of DAO, i.e., the electron-proton-electron transfer
mechanism and the ionic mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Katane,
Y.Saitoh,
K.Maeda,
T.Hanai,
M.Sekine,
T.Furuchi,
and
H.Homma
(2011).
Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian D-aspartate oxidase.
|
| |
Amino Acids,
40,
467-476.
|
 |
|
|
|
|
 |
M.Katane,
and
H.Homma
(2010).
D-aspartate oxidase: the sole catabolic enzyme acting on free D-aspartate in mammals.
|
| |
Chem Biodivers,
7,
1435-1449.
|
 |
|
|
|
|
 |
C.J.Carrell,
R.C.Bruckner,
D.Venci,
G.Zhao,
M.S.Jorns,
and
F.S.Mathews
(2007).
NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution.
|
| |
Structure,
15,
928-941.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bakke,
C.Setoyama,
R.Miura,
and
N.Kajiyama
(2006).
Thermostabilization of porcine kidney D-amino acid oxidase by a single amino acid substitution.
|
| |
Biotechnol Bioeng,
93,
1023-1027.
|
 |
|
|
|
|
 |
T.Kawazoe,
H.Tsuge,
M.S.Pilone,
and
K.Fukui
(2006).
Crystal structure of human D-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch located at the si-face of the flavin ring.
|
| |
Protein Sci,
15,
2708-2717.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Binda,
R.Angelini,
R.Federico,
P.Ascenzi,
and
A.Mattevi
(2001).
Structural bases for inhibitor binding and catalysis in polyamine oxidase.
|
| |
Biochemistry,
40,
2766-2776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Yurimoto,
T.Hasegawa,
Y.Sakai,
and
N.Kato
(2000).
Physiological role of the D-amino acid oxidase gene, DAO1, in carbon and nitrogen metabolism in the methylotrophic yeast Candida boidinii.
|
| |
Yeast,
16,
1217-1227.
|
 |
|
|
|
|
 |
P.D.Pawelek,
J.Cheah,
R.Coulombe,
P.Macheroux,
S.Ghisla,
and
A.Vrielink
(2000).
The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
|
| |
EMBO J,
19,
4204-4215.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Umhau,
L.Pollegioni,
G.Molla,
K.Diederichs,
W.Welte,
M.S.Pilone,
and
S.Ghisla
(2000).
The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
|
| |
Proc Natl Acad Sci U S A,
97,
12463-12468.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Trickey,
M.A.Wagner,
M.S.Jorns,
and
F.S.Mathews
(1999).
Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme.
|
| |
Structure,
7,
331-345.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

