 |
PDBsum entry 1aa8
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.3
- D-amino-acid oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Cephalosporin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a D-alpha-amino acid + O2 + H2O = a 2-oxocarboxylate + H2O2 + NH4+
|
 |
 |
 |
 |
 |
D-alpha-amino acid
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
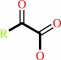 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
matches with 41.33% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
120:14-17
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 A resolution.
|
|
H.Mizutani,
I.Miyahara,
K.Hirotsu,
Y.Nishina,
K.Shiga,
C.Setoyama,
R.Miura.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystallographic structure of porcine kidney D-amino acid oxidase,
which had been expressed in Escherichia coli transformed with a vector
containing DAO cDNA, was determined by the isomorphous replacement method for
the complex form with benzoate. The known amino acid sequence, FAD and benzoate
were fitted to an electron density map of 3.0 A resolution with an R-factor of
21.0%. The overall dimeric structure exhibits an elongated ellipsoidal
framework. The prosthetic group, FAD, was found to be in an extended
conformation, the isoalloxazine ring being buried in the protein core. The ADP
moiety of FAD was located in the typical beta alpha beta dinucleotide binding
motif, with the alpha-helix dipole stabilizing the pyrophosphate negative
charge. The substrate analog, benzoate, is located on the re-face of the
isoalloxazine ring, while the si-face is blocked by hydrophobic residues. The
carboxylate group of benzoate is ion-paired with the Arg283 side chain and is
within interacting distance with the hydroxy moiety of Tyr228. The phenol ring
of Tyr224 is located just above the benzene ring of benzoate, implying the
importance of this residue for catalysis. There is no positive charge or
alpha-helix dipole near N(1) of flavin. Hydrogen bonds were observed at C(2) =
O, N(3)-H, C(4) = O, and N(5) of the flavin ring.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Katane,
Y.Saitoh,
K.Maeda,
T.Hanai,
M.Sekine,
T.Furuchi,
and
H.Homma
(2011).
Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian D-aspartate oxidase.
|
| |
Amino Acids,
40,
467-476.
|
 |
|
|
|
|
 |
H.Soetedjo,
M.F.Mora,
and
C.D.Garcia
(2010).
Optical Properties of Single-Wall Carbon Nanotube Films Deposited on Si/SiO(2) Wafers.
|
| |
Thin Solid Films,
518,
3954-3959.
|
 |
|
|
|
|
 |
L.Pollegioni,
and
S.Sacchi
(2010).
Metabolism of the neuromodulator D-serine.
|
| |
Cell Mol Life Sci,
67,
2387-2404.
|
 |
|
|
|
|
 |
M.Katane,
and
H.Homma
(2010).
D-aspartate oxidase: the sole catabolic enzyme acting on free D-aspartate in mammals.
|
| |
Chem Biodivers,
7,
1435-1449.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2010).
Oxidation of amines by flavoproteins.
|
| |
Arch Biochem Biophys,
493,
13-25.
|
 |
|
|
|
|
 |
A.Gholizadeh,
and
B.B.Kohnehrouz
(2009).
Molecular cloning and expression in Escherichia coli of an active fused Zea mays L. D-amino acid oxidase.
|
| |
Biochemistry (Mosc),
74,
137-144.
|
 |
|
|
|
|
 |
M.F.Mora,
C.E.Giacomelli,
and
C.D.Garcia
(2009).
Interaction of D-amino acid oxidase with carbon nanotubes: implications in the design of biosensors.
|
| |
Anal Chem,
81,
1016-1022.
|
 |
|
|
|
|
 |
M.Bakke,
C.Setoyama,
R.Miura,
and
N.Kajiyama
(2006).
Thermostabilization of porcine kidney D-amino acid oxidase by a single amino acid substitution.
|
| |
Biotechnol Bioeng,
93,
1023-1027.
|
 |
|
|
|
|
 |
T.Kawazoe,
H.Tsuge,
M.S.Pilone,
and
K.Fukui
(2006).
Crystal structure of human D-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch located at the si-face of the flavin ring.
|
| |
Protein Sci,
15,
2708-2717.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Koch,
C.Breithaupt,
R.Kiefersauer,
J.Freigang,
R.Huber,
and
A.Messerschmidt
(2004).
Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
|
| |
EMBO J,
23,
1720-1728.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Wille,
M.Ritter,
R.Friedemann,
W.Mäntele,
and
G.Hübner
(2003).
Redox-triggered FTIR difference spectra of FAD in aqueous solution and bound to flavoproteins.
|
| |
Biochemistry,
42,
14814-14821.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
R.Miura
(2001).
Versatility and specificity in flavoenzymes: control mechanisms of flavin reactivity.
|
| |
Chem Rec,
1,
183-194.
|
 |
|
|
|
|
 |
Y.Liu,
T.M.Louie,
J.Payne,
J.Bohuslavek,
H.Bolton,
and
L.Xun
(2001).
Identification, purification, and characterization of iminodiacetate oxidase from the EDTA-degrading bacterium BNC1.
|
| |
Appl Environ Microbiol,
67,
696-701.
|
 |
|
|
|
|
 |
A.A.Raibekas,
K.Fukui,
and
V.Massey
(2000).
Design and properties of human D-amino acid oxidase with covalently attached flavin.
|
| |
Proc Natl Acad Sci U S A,
97,
3089-3093.
|
 |
|
|
|
|
 |
E.Varela,
M.Jesús Martínez,
and
A.T.Martínez
(2000).
Aryl-alcohol oxidase protein sequence: a comparison with glucose oxidase and other FAD oxidoreductases.
|
| |
Biochim Biophys Acta,
1481,
202-208.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Yorita,
H.Misaki,
B.A.Palfey,
and
V.Massey
(2000).
On the interpretation of quantitative structure-function activity relationship data for lactate oxidase.
|
| |
Proc Natl Acad Sci U S A,
97,
2480-2485.
|
 |
|
|
|
|
 |
P.D.Pawelek,
J.Cheah,
R.Coulombe,
P.Macheroux,
S.Ghisla,
and
A.Vrielink
(2000).
The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
|
| |
EMBO J,
19,
4204-4215.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Umhau,
L.Pollegioni,
G.Molla,
K.Diederichs,
W.Welte,
M.S.Pilone,
and
S.Ghisla
(2000).
The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
|
| |
Proc Natl Acad Sci U S A,
97,
12463-12468.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Mattevi,
G.Tedeschi,
L.Bacchella,
A.Coda,
A.Negri,
and
S.Ronchi
(1999).
Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family.
|
| |
Structure,
7,
745-756.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Gabler,
and
L.Fischer
(1999).
Production of a new D-amino acid oxidase from the fungus Fusarium oxysporum.
|
| |
Appl Environ Microbiol,
65,
3750-3753.
|
 |
|
|
|
|
 |
P.Trickey,
M.A.Wagner,
M.S.Jorns,
and
F.S.Mathews
(1999).
Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme.
|
| |
Structure,
7,
331-345.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Mattevi
(1998).
The PHBH fold: not only flavoenzymes.
|
| |
Biophys Chem,
70,
217-222.
|
 |
|
|
|
|
 |
K.S.Rao,
and
F.Lederer
(1998).
About the pKa of the active-site histidine in flavocytochrome b2 (yeast L-lactate dehydrogenase).
|
| |
Protein Sci,
7,
1531-1537.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

