 |
PDBsum entry 1a59

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Cold-activity
|
PDB id
|
|

|

|
1a59
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.3.3.16
- citrate synthase (unknown stereospecificity).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxaloacetate + acetyl-CoA + H2O = citrate + CoA + H+
|
 |
 |
 |
 |
 |
 oxaloacetate
oxaloacetate
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
H2O
|
=
|
citrate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
CoA
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.3.3.5
- 2-methylcitrate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
propanoyl-CoA + oxaloacetate + H2O = (2S,3S)-2-methylcitrate + CoA + H+
|
 |
 |
 |
 |
 |
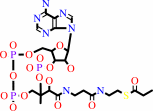 propanoyl-CoA
propanoyl-CoA
|
+
|
oxaloacetate
Bound ligand (Het Group name = )
matches with 69.23% similarity
|
+
|
H2O
|
=
|
(2S,3S)-2-methylcitrate
|
+
|
CoA
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
6:351-361
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium.
|
|
R.J.Russell,
U.Gerike,
M.J.Danson,
D.W.Hough,
G.L.Taylor.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The structural basis of adaptation of enzymes to low temperature is
poorly understood. Dimeric citrate synthase has been used as a model enzyme to
study the structural basis of thermostability, the structure of the enzyme from
organisms living in habitats at 55 degrees C and 100 degrees C having previously
been determined. Here the study is extended to include a citrate synthase from
an Antarctic bacterium, allowing us to explore the structural basis of cold
activity and thermostability across the whole temperature range over which life
is known to exit. RESULTS: We report here the first crystal structure of a
cold-active enzyme, citrate synthase, isolated from an Antarctic bacterium, at a
resolution of 2.09 A. In comparison with the same enzyme from a
hyperthermophilic host, the cold-active enzyme has a much more accessible active
site, an unusual electrostatic potential distribution and an increased relative
flexibility of the small domain compared to the large domain. Several other
features of the cold-active enzyme were also identified: reduced subunit
interface interactions with no intersubunit ion-pair networks; loops of
increased length carrying more charge and fewer proline residues; an increase in
solvent-exposed hydrophobic residues; and an increase in intramolecular ion
pairs. CONCLUSIONS: Enzymes from organisms living at the temperature extremes of
life need to avoid hot or cold denaturation yet maintain sufficient structural
integrity to allow catalytic efficiency. For hyperthermophiles, thermal
denaturation of the citrate synthase dimer appears to be resisted by complex
networks of ion pairs at the dimer interface, a feature common to other
hyperthermophilic proteins. For the cold-active citrate synthase, cold
denaturation appears to be resisted by an increase in intramolecular ion pairs
compared to the hyperthermophilic enzyme. Catalytic efficiency of the
cold-active enzyme appears to be achieved by a more accessible active site and
by an increase in the relative flexibility of the small domain compared to the
large domain.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1998,
6,
351-361)
copyright 1998.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Chittori,
H.S.Savithri,
and
M.R.Murthy
(2011).
Crystal structure of Salmonella typhimurium 2-methylcitrate synthase: Insights on domain movement and substrate specificity.
|
| |
J Struct Biol,
174,
58-68.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Prakash,
and
N.Jaiswal
(2010).
alpha-Amylase: an ideal representative of thermostable enzymes.
|
| |
Appl Biochem Biotechnol,
160,
2401-2414.
|
 |
|
|
|
|
 |
R.M.Evans,
E.M.Behiry,
L.H.Tey,
J.Guo,
E.J.Loveridge,
and
R.K.Allemann
(2010).
Catalysis by dihydrofolate reductase from the psychropiezophile Moritella profunda.
|
| |
Chembiochem,
11,
2010-2017.
|
 |
|
|
|
|
 |
B.B.Xie,
F.Bian,
X.L.Chen,
H.L.He,
J.Guo,
X.Gao,
Y.X.Zeng,
B.Chen,
B.C.Zhou,
and
Y.Z.Zhang
(2009).
Cold adaptation of zinc metalloproteases in the thermolysin family from deep sea and arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics: new insights into relationship between conformational flexibility and hydrogen bonding.
|
| |
J Biol Chem,
284,
9257-9269.
|
 |
|
|
|
|
 |
Y.T.Aminetzach,
J.R.Srouji,
C.Y.Kong,
and
H.E.Hoekstra
(2009).
Convergent evolution of novel protein function in shrew and lizard venom.
|
| |
Curr Biol,
19,
1925-1931.
|
 |
|
|
|
|
 |
C.Bauvois,
L.Jacquamet,
A.L.Huston,
F.Borel,
G.Feller,
and
J.L.Ferrer
(2008).
Crystal structure of the cold-active aminopeptidase from Colwellia psychrerythraea, a close structural homologue of the human bifunctional leukotriene A4 hydrolase.
|
| |
J Biol Chem,
283,
23315-23325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.F.Rodrigues,
and
J.M.Tiedje
(2008).
Coping with our cold planet.
|
| |
Appl Environ Microbiol,
74,
1677-1686.
|
 |
|
|
|
|
 |
M.Olufsen,
A.O.Smalås,
and
B.O.Brandsdal
(2008).
Electrostatic interactions play an essential role in DNA repair and cold-adaptation of Uracil DNA glycosylase.
|
| |
J Mol Model,
14,
201-213.
|
 |
|
|
|
|
 |
M.Olufsen,
E.Papaleo,
A.O.Smalås,
and
B.O.Brandsdal
(2008).
Ion pairs and their role in modulating stability of cold- and warm-active uracil DNA glycosylase.
|
| |
Proteins,
71,
1219-1230.
|
 |
|
|
|
|
 |
W.C.Too,
Y.C.Liew,
and
L.L.Few
(2008).
Cloning of glyceraldehyde-3-phosphate dehydrogenase from an Antarctic psychrophilic bacterium by inverse and splinkerette PCR.
|
| |
J Basic Microbiol,
48,
430-435.
|
 |
|
|
|
|
 |
D.R.Boutz,
D.Cascio,
J.Whitelegge,
L.J.Perry,
and
T.O.Yeates
(2007).
Discovery of a thermophilic protein complex stabilized by topologically interlinked chains.
|
| |
J Mol Biol,
368,
1332-1344.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Tronelli,
E.Maugini,
F.Bossa,
and
S.Pascarella
(2007).
Structural adaptation to low temperatures--analysis of the subunit interface of oligomeric psychrophilic enzymes.
|
| |
FEBS J,
274,
4595-4608.
|
 |
|
|
|
|
 |
E.K.Riise,
M.S.Lorentzen,
R.Helland,
A.O.Smalås,
H.K.Leiros,
and
N.P.Willassen
(2007).
The first structure of a cold-active catalase from Vibrio salmonicida at 1.96 A reveals structural aspects of cold adaptation.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
135-148.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Hårdeman,
and
S.Sjöling
(2007).
Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment.
|
| |
FEMS Microbiol Ecol,
59,
524-534.
|
 |
|
|
|
|
 |
G.S.Garvey,
C.J.Rocco,
J.C.Escalante-Semerena,
and
I.Rayment
(2007).
The three-dimensional crystal structure of the PrpF protein of Shewanella oneidensis complexed with trans-aconitate: insights into its biological function.
|
| |
Protein Sci,
16,
1274-1284.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Spiwok,
P.Lipovová,
T.Skálová,
J.Dusková,
J.Dohnálek,
J.Hasek,
N.J.Russell,
and
B.Králová
(2007).
Cold-active enzymes studied by comparative molecular dynamics simulation.
|
| |
J Mol Model,
13,
485-497.
|
 |
|
|
|
|
 |
J.A.Coker,
and
J.E.Brenchley
(2006).
Protein engineering of a cold-active beta-galactosidase from Arthrobacter sp. SB to increase lactose hydrolysis reveals new sites affecting low temperature activity.
|
| |
Extremophiles,
10,
515-524.
|
 |
|
|
|
|
 |
K.S.Siddiqui,
and
R.Cavicchioli
(2006).
Cold-adapted enzymes.
|
| |
Annu Rev Biochem,
75,
403-433.
|
 |
|
|
|
|
 |
O.A.Adekoya,
R.Helland,
N.P.Willassen,
and
I.Sylte
(2006).
Comparative sequence and structure analysis reveal features of cold adaptation of an enzyme in the thermolysin family.
|
| |
Proteins,
62,
435-449.
|
 |
|
|
|
|
 |
A.Seto,
K.Murayama,
M.Toyama,
A.Ebihara,
N.Nakagawa,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2005).
ATP-induced structural change of dephosphocoenzyme A kinase from Thermus thermophilus HB8.
|
| |
Proteins,
58,
235-242.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Dong,
T.Ihara,
H.Motoshima,
and
K.Watanabe
(2005).
Crystallization and preliminary X-ray crystallographic studies of a psychrophilic subtilisin-like protease Apa1 from Antarctic Pseudoalteromonas sp. strain AS-11.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
308-311.
|
 |
|
|
|
|
 |
J.Arnórsdóttir,
M.M.Kristjánsson,
and
R.Ficner
(2005).
Crystal structure of a subtilisin-like serine proteinase from a psychrotrophic Vibrio species reveals structural aspects of cold adaptation.
|
| |
FEBS J,
272,
832-845.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Olufsen,
A.O.Smalås,
E.Moe,
and
B.O.Brandsdal
(2005).
Increased flexibility as a strategy for cold adaptation: a comparative molecular dynamics study of cold- and warm-active uracil DNA glycosylase.
|
| |
J Biol Chem,
280,
18042-18048.
|
 |
|
|
|
|
 |
A.Hoyoux,
V.Blaise,
T.Collins,
S.D'Amico,
E.Gratia,
A.L.Huston,
J.C.Marx,
G.Sonan,
Y.Zeng,
G.Feller,
and
C.Gerday
(2004).
Extreme catalysts from low-temperature environments.
|
| |
J Biosci Bioeng,
98,
317-330.
|
 |
|
|
|
|
 |
D.Georlette,
V.Blaise,
T.Collins,
S.D'Amico,
E.Gratia,
A.Hoyoux,
J.C.Marx,
G.Sonan,
G.Feller,
and
C.Gerday
(2004).
Some like it cold: biocatalysis at low temperatures.
|
| |
FEMS Microbiol Rev,
28,
25-42.
|
 |
|
|
|
|
 |
H.Tsuruta,
J.Tamura,
H.Yamagata,
and
Y.Aizono
(2004).
Specification of amino acid residues essential for the catalytic reaction of cold-active protein-tyrosine phosphatase of a psychrophile, Shewanella sp.
|
| |
Biosci Biotechnol Biochem,
68,
440-443.
|
 |
|
|
|
|
 |
S.Kumar,
and
R.Nussinov
(2004).
Different roles of electrostatics in heat and in cold: adaptation by citrate synthase.
|
| |
Chembiochem,
5,
280-290.
|
 |
|
|
|
|
 |
F.Van Petegem,
T.Collins,
M.A.Meuwis,
C.Gerday,
G.Feller,
and
J.Van Beeumen
(2003).
The structure of a cold-adapted family 8 xylanase at 1.3 A resolution. Structural adaptations to cold and investgation of the active site.
|
| |
J Biol Chem,
278,
7531-7539.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Feller,
and
C.Gerday
(2003).
Psychrophilic enzymes: hot topics in cold adaptation.
|
| |
Nat Rev Microbiol,
1,
200-208.
|
 |
|
|
|
|
 |
I.Leiros,
E.Moe,
O.Lanes,
A.O.Smalås,
and
N.P.Willassen
(2003).
The structure of uracil-DNA glycosylase from Atlantic cod (Gadus morhua) reveals cold-adaptation features.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1357-1365.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Aghajari,
F.Van Petegem,
V.Villeret,
J.P.Chessa,
C.Gerday,
R.Haser,
and
J.Van Beeumen
(2003).
Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold-adapted proteases.
|
| |
Proteins,
50,
636-647.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Brautaset,
M.D.Williams,
R.D.Dillingham,
C.Kaufmann,
A.Bennaars,
E.Crabbe,
and
M.C.Flickinger
(2003).
Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in L-lysine-secreting mutants.
|
| |
Appl Environ Microbiol,
69,
3986-3995.
|
 |
|
|
|
|
 |
G.Gianese,
F.Bossa,
and
S.Pascarella
(2002).
Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes.
|
| |
Proteins,
47,
236-249.
|
 |
|
|
|
|
 |
G.Ko,
M.W.First,
and
H.A.Burge
(2002).
The characterization of upper-room ultraviolet germicidal irradiation in inactivating airborne microorganisms.
|
| |
Environ Health Perspect,
110,
95.
|
 |
|
|
|
|
 |
G.S.Bell,
R.J.Russell,
H.Connaris,
D.W.Hough,
M.J.Danson,
and
G.L.Taylor
(2002).
Stepwise adaptations of citrate synthase to survival at life's extremes. From psychrophile to hyperthermophile.
|
| |
Eur J Biochem,
269,
6250-6260.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Arnórsdottir,
R.B.Smáradóttir,
O.T.Magnússon,
S.H.Thorbjarnardóttir,
G.Eggertsson,
and
M.M.Kristjánsson
(2002).
Characterization of a cloned subtilisin-like serine proteinase from a psychrotrophic Vibrio species.
|
| |
Eur J Biochem,
269,
5536-5546.
|
 |
|
|
|
|
 |
S.D'Amico,
P.Claverie,
T.Collins,
D.Georlette,
E.Gratia,
A.Hoyoux,
M.A.Meuwis,
G.Feller,
and
C.Gerday
(2002).
Molecular basis of cold adaptation.
|
| |
Philos Trans R Soc Lond B Biol Sci,
357,
917-925.
|
 |
|
|
|
|
 |
F.H.Arnold,
P.L.Wintrode,
K.Miyazaki,
and
A.Gershenson
(2001).
How enzymes adapt: lessons from directed evolution.
|
| |
Trends Biochem Sci,
26,
100-106.
|
 |
|
|
|
|
 |
I.Tsigos,
K.Mavromatis,
M.Tzanodaskalaki,
C.Pozidis,
M.Kokkinidis,
and
V.Bouriotis
(2001).
Engineering the properties of a cold active enzyme through rational redesign of the active site.
|
| |
Eur J Biochem,
268,
5074-5080.
|
 |
|
|
|
|
 |
T.Lonhienne,
K.Mavromatis,
C.E.Vorgias,
L.Buchon,
C.Gerday,
and
V.Bouriotis
(2001).
Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes.
|
| |
J Bacteriol,
183,
1773-1779.
|
 |
|
|
|
|
 |
W.Dzwolak,
M.Kato,
A.Shimizu,
and
Y.Taniguchi
(2001).
FTIR study on heat-induced and pressure-assisted cold-induced changes in structure of bovine alpha-lactalbumin: stabilizing role of calcium ion.
|
| |
Biopolymers,
62,
29-39.
|
 |
|
|
|
|
 |
C.Gerday,
M.Aittaleb,
M.Bentahir,
J.P.Chessa,
P.Claverie,
T.Collins,
S.D'Amico,
J.Dumont,
G.Garsoux,
D.Georlette,
A.Hoyoux,
T.Lonhienne,
M.A.Meuwis,
and
G.Feller
(2000).
Cold-adapted enzymes: from fundamentals to biotechnology.
|
| |
Trends Biotechnol,
18,
103-107.
|
 |
|
|
|
|
 |
D.Georlette,
Z.O.Jónsson,
F.Van Petegem,
J.Chessa,
J.Van Beeumen,
U.Hübscher,
and
C.Gerday
(2000).
A DNA ligase from the psychrophile Pseudoalteromonas haloplanktis gives insights into the adaptation of proteins to low temperatures.
|
| |
Eur J Biochem,
267,
3502-3512.
|
 |
|
|
|
|
 |
H.K.Leiros,
N.P.Willassen,
and
A.O.Smalås
(2000).
Structural comparison of psychrophilic and mesophilic trypsins. Elucidating the molecular basis of cold-adaptation.
|
| |
Eur J Biochem,
267,
1039-1049.
|
 |
|
|
|
|
 |
L.C.Kurz,
G.Drysdale,
M.Riley,
M.A.Tomar,
J.Chen,
R.J.Russell,
and
M.J.Danson
(2000).
Kinetics and mechanism of the citrate synthase from the thermophilic archaeon Thermoplasma acidophilum.
|
| |
Biochemistry,
39,
2283-2296.
|
 |
|
|
|
|
 |
M.Rina,
C.Pozidis,
K.Mavromatis,
M.Tzanodaskalaki,
M.Kokkinidis,
and
V.Bouriotis
(2000).
Alkaline phosphatase from the Antarctic strain TAB5. Properties and psychrophilic adaptations.
|
| |
Eur J Biochem,
267,
1230-1238.
|
 |
|
|
|
|
 |
N.Panasik,
J.E.Brenchley,
and
G.K.Farber
(2000).
Distributions of structural features contributing to thermostability in mesophilic and thermophilic alpha/beta barrel glycosyl hydrolases.
|
| |
Biochim Biophys Acta,
1543,
189-201.
|
 |
|
|
|
|
 |
T.Lonhienne,
C.Gerday,
and
G.Feller
(2000).
Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility.
|
| |
Biochim Biophys Acta,
1543,
1.
|
 |
|
|
|
|
 |
Y.Okubo,
K.Yokoigawa,
N.Esaki,
K.Soda,
and
H.Misono
(2000).
High catalytic activity of alanine racemase from psychrophilic Bacillus psychrosaccharolyticus at high temperatures in the presence of pyridoxal 5'-phosphate.
|
| |
FEMS Microbiol Lett,
192,
169-173.
|
 |
|
|
|
|
 |
A.Ayed,
and
H.W.Duckworth
(1999).
A stable intermediate in the equilibrium unfolding of Escherichia coli citrate synthase.
|
| |
Protein Sci,
8,
1116-1126.
|
 |
|
|
|
|
 |
A.Galkin,
L.Kulakova,
H.Ashida,
Y.Sawa,
and
N.Esaki
(1999).
Cold-adapted alanine dehydrogenases from two antarctic bacterial strains: gene cloning, protein characterization, and comparison with mesophilic and thermophilic counterparts.
|
| |
Appl Environ Microbiol,
65,
4014-4020.
|
 |
|
|
|
|
 |
D.Maes,
J.P.Zeelen,
N.Thanki,
N.Beaucamp,
M.Alvarez,
M.H.Thi,
J.Backmann,
J.A.Martial,
L.Wyns,
R.Jaenicke,
and
R.K.Wierenga
(1999).
The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures.
|
| |
Proteins,
37,
441-453.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.W.Hough,
and
M.J.Danson
(1999).
Extremozymes.
|
| |
Curr Opin Chem Biol,
3,
39-46.
|
 |
|
|
|
|
 |
S.Y.Kim,
K.Y.Hwang,
S.H.Kim,
H.C.Sung,
Y.S.Han,
and
Y.Cho
(1999).
Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
|
| |
J Biol Chem,
274,
11761-11767.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Danson,
and
D.W.Hough
(1998).
Structure, function and stability of enzymes from the Archaea.
|
| |
Trends Microbiol,
6,
307-314.
|
 |
|
|
|
|
 |
N.Aghajari,
G.Feller,
C.Gerday,
and
R.Haser
(1998).
Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level.
|
| |
Structure,
6,
1503-1516.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

