 |
PDBsum entry 9j9s
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Artificial mononuclear zn-bound metalloprotein 5 (m5:zn)
|
|
Structure:
|
 |
Dtdp-4-dehydrorhamnose 3,5-epimerase. Chain: a, b, c, d. Synonym: thymidine diphospho-4-keto-rhamnose 3,5-epimerase,dtdp-4- keto-6-deoxyglucose 3,dtdp-6-deoxy-d-xylo-4-hexulose 3,dtdp-l- rhamnose synthase. Engineered: yes. Other_details: deoxythymidine diphosphate (dtdp)-4-keto-6-deoxy-d- hexulose 3, 5-epimerase (rmlc)
|
|
Source:
|
 |
Methanothermobacter thermautotrophicus. Organism_taxid: 145262. Gene: rmlc, mth_1790. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.259
|
R-free:
|
0.305
|
|
|
Authors:
|
 |
W.J.Jeong,W.J.Song
|
|
Key ref:
|
 |
W.J.Jeong
and
w.j.song
Metal-Installer: a protein designer tool to create metal-Binding sites.
To be published,
.
|
 |
|
Date:
|
 |
|
23-Aug-24
|
Release date:
|
02-Jul-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O27818
(RMLC_METTH) -
dTDP-4-dehydrorhamnose 3,5-epimerase from Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
185 a.a.
183 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.3.13
- dTDP-4-dehydrorhamnose 3,5-epimerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
6-Deoxyhexose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dTDP-4-dehydro-6-deoxy-alpha-D-glucose = dTDP-4-dehydro-beta-L-rhamnose
|
 |
 |
 |
 |
 |
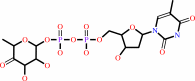 dTDP-4-dehydro-6-deoxy-alpha-D-glucose
dTDP-4-dehydro-6-deoxy-alpha-D-glucose
|
=
|
dTDP-4-dehydro-beta-L-rhamnose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
NAD(+)
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

