 |
PDBsum entry 9fxh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of cobalt(ii)-substituted double mutant y115e y117e human glutaminyl cyclase
|
|
Structure:
|
 |
Glutaminyl-peptide cyclotransferase. Chain: a, b, c. Synonym: glutaminyl cyclase,qc,sqc,glutaminyl-tRNA cyclotransferase, glutamyl cyclase,ec. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: qpct. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.207
|
R-free:
|
0.257
|
|
|
Authors:
|
 |
G.Tassone,C.Pozzi,S.Mangani
|
|
Key ref:
|
 |
G.Tassone
et al.
(2024).
Metal ion binding to human glutaminyl cyclase: a stru perspective..
Int j mol sci,
25,
.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Jul-24
|
Release date:
|
04-Sep-24
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16769
(QPCT_HUMAN) -
Glutaminyl-peptide cyclotransferase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
361 a.a.
323 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.2.5
- glutaminyl-peptide cyclotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-terminal L-glutaminyl-[peptide] = N-terminal 5-oxo-L-prolyl-[peptide] + NH4+
|
 |
 |
 |
 |
 |
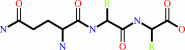 L-glutaminyl-peptide
L-glutaminyl-peptide
|
=
|
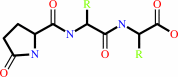 5-oxoprolyl-peptide
5-oxoprolyl-peptide
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

