 |
PDBsum entry 8ygc

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 983 a.a.
983 a.a.
|
 |

|

|

|

|

|

|

|

 151 a.a.
151 a.a.
|
 |

|

|

|

|

|

|

|

 277 a.a.
277 a.a.
|
 |

|

|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
The dimer structure of dsr2-spr
|
|
Structure:
|
 |
Sir2-like domain-containing protein. Chain: a, b, e, f. Engineered: yes. Mutation: yes. Other_details: sequence reference for bacillus subtilis a29 (2508883) is not available in uniprot at the time of biocuration. Current sequence reference is from uniprot id d4g637.. Spr. Chain: c, d.
|
|
Source:
|
 |
Bacillus subtilis a29. Organism_taxid: 2508883. Gene: bsnt_07056. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: b4122_1986, j5227_09585. Expression_system_taxid: 562
|
|
Authors:
|
 |
X.Gao,H.Zhu,S.Cui
|
|
Key ref:
|
 |
K.Zhu
et al.
(2025).
Activation of the bacterial defense-Associated sirtui system..
Commun biol,
8,
297.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
26-Feb-24
|
Release date:
|
05-Mar-25
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0DXN8
(DSR2_BACIU) -
Defense-associated sirtuin 2 from Bacillus subtilis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1005 a.a.
983 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, E, F:
E.C.3.2.2.5
- NAD(+) glycohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
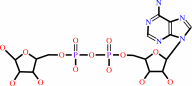 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
| |

