 |
PDBsum entry 8ubc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
RNA binding protein/RNA
|
PDB id
|
|

|

|
8ubc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 315 a.a.
315 a.a.
|
 |

|

|

|

|

|

|

|
 122 a.a.
122 a.a.
|
 |

|

|

|

|

|

|

|



 111 a.a.
111 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
RNA binding protein/RNA
|
 |
|
Title:
|
 |
Diversity-generating retroelement (dgr) ribonucleoprotein reverse transcriptase - resting state 1b
|
|
Structure:
|
 |
Reverse transcriptase. Chain: a. Engineered: yes. Avd. Chain: b, c, d, e, f. Synonym: formaldehyde-activating enzyme,fae. Engineered: yes. Diversity-generating retroelement (dgr) RNA tr. Chain: h.
|
|
Source:
|
 |
Bordetella phage bpp-1. Organism_taxid: 2885909. Gene: brt. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: bbp7, fae, mexam1_meta1p1766. Synthetic: yes. Organism_taxid: 2885909
|
|
Authors:
|
 |
T.Biswas,S.Handa,P.Ghosh
|
|
Key ref:
|
 |
S.Handa
et al.
Rna control of reverse transcription in a diversity-Generating retroelement.
Nature,
.
PubMed id:
|
 |
|
Date:
|
 |
|
22-Sep-23
|
Release date:
|
23-Oct-24
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q775D8
(Q775D8_BPBPP) -
Reverse transcriptase from Bordetella phage BPP-1
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
328 a.a.
315 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q775D7
(Q775D7_BPBPP) -
Bbp7 from Bordetella phage BPP-1
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
128 a.a.
122 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q9FA38
(FAE_METEA) -
5,6,7,8-tetrahydromethanopterin hydro-lyase from Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
170 a.a.
122 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains B, C, D, E, F:
E.C.4.2.1.147
- 5,6,7,8-tetrahydromethanopterin hydro-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5,6,7,8-tetrahydromethanopterin + formaldehyde = 5,10- methylenetetrahydromethanopterin + H2O
|
 |
 |
 |
 |
 |
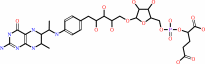 5,6,7,8-tetrahydromethanopterin
5,6,7,8-tetrahydromethanopterin
|
+
|
 formaldehyde
formaldehyde
|
=
|
 5,10- methylenetetrahydromethanopterin
5,10- methylenetetrahydromethanopterin
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |

| | |

