 |
PDBsum entry 8h2a

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
8h2a
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of alcohol dehydrogenase from formosa agariphila
|
|
Structure:
|
 |
Alcohol dehydrogenase. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Formosa agariphila. Organism_taxid: 320324. Strain: dsm 15362 / kctc 12365 / lmg 23005 / kmm 3901 / m-2alg 35-1. Gene: bn863_21030. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.226
|
R-free:
|
0.278
|
|
|
Authors:
|
 |
S.Brott,U.T.Bornscheuer,K.H.Nam
|
|
Key ref:
|
 |
S.Brott
et al.
Crystal structure of alcohol dehydrogenase from formo agariphila.
To be published,
.
|
 |
|
Date:
|
 |
|
05-Oct-22
|
Release date:
|
18-Oct-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
T2KM87
(T2KM87_9FLAO) -
Alcohol dehydrogenase from Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
370 a.a.
366 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
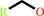 primary alcohol
primary alcohol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
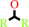 secondary alcohol
secondary alcohol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
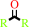 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

